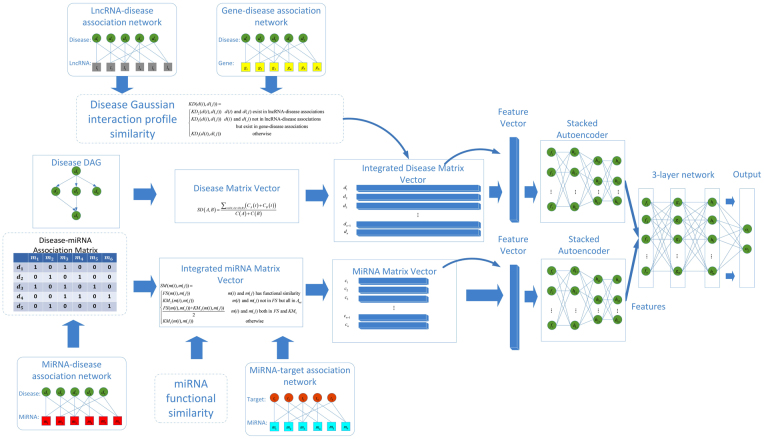
Figure 3.
The flowchart of proposed DeepMDA. The miRNA similarity was integrated using miRNA-functional similarity and miRNA-disease association. As for disease similarity, we adopted DAG information and Gaussian interaction profile similarity information. The two input data was fed into two stacked autoencoders to learn high-level features, then merged and finally utilized a 3-layer network to infer the association between miRNAs and diseases.