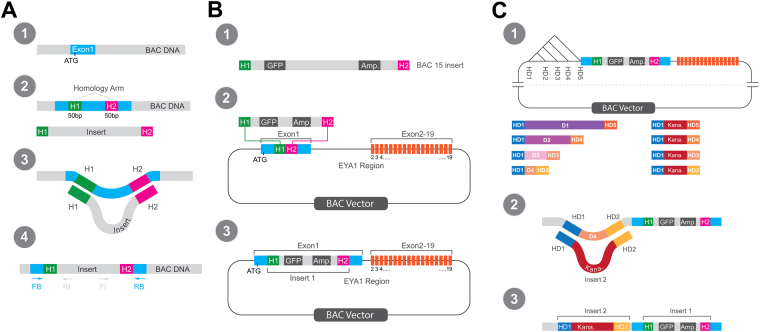
Figure 3.
Double recombineering strategy to localize enhancers in large genomic regions. (A) Overview of recombineering procedure. A target region for insertion is selected in the BAC, for example the first coding exon of the gene of interest containing the start codon (ATG) (1). An insert (containing an antibiotic resistance gene and possibly other elements such as a reporter gene) with appropriate homology arms (H1, H2) complementary to BAC target sequences is transformed into bacteria containing the BAC (2). The insert is inserted into the BAC by homologous recombination (recombineering) replacing the original BAC sequence between H1 and H2 (3). Proper insertion is verified by PCR using forward (FB) and reverse primers (RB) for BAC sequences flanking the insertion, while proper orientation is verified by PCR combining FB with a reverse primer for an insert specific sequence (RI) or a forward primer for an insert specific sequence with RB (4). (B) Creation of reporter BACs in the first recombineering step. Insert 1 (here the insert for BAC15) contains an ampicillin resistance gene and a GFP reporter gene (1). Homology arms are designed to target the insert to a position immediately after the start codon in the first coding exon of Eya1 during recombineering (2). BAC reporter construct after recombineering (3). (C) Creation of deletion constructs of BAC reporters in the second recombineering step. Sequences flanking large noncoding regions in the BAC are selected as new homology arms (HD1-HD5). Chosing different combinations of these for the insert 2 (containing a kanamycin resistance gene) will result in deletions of different extent (1). During recombineering the sequence D4 on the BAC between HD1 and HD2 (or between other homology arms chosen) will be deleted and replaced by the kanamycin resistance cassette (2). BAC reporter deletion construct after second step of recombineering containing insert 1 and 2 (3).