Abstract
The non-expressor of the pathogenesis-related genes 1 (NPR1) is a master regulator in defense signaling of plants and plays a key role in basal and systemic acquired resistance. In this study, we isolated a NPR1-like gene from the oriental hybrid lily ‘Sorbonne’ (designated as LhSorNPR1) using rapid amplification of cDNA ends (RACE). The open reading frame of LhSorNPR1 consisted of 1854 bp, encoding a protein of 617 amino acids. Multiple sequence alignment revealed that LhSorNPR1 shares high similarity to NPR1-like proteins and characteristics of the BTB/POZ domain and ankyrin repeats. A comparison between the intron/exon organization of LhSorNPR1 and orthologs from other plant species demonstrated that NPR1 genomic fragments (including LhSorNPR1) are all composed of 4 exons and 3 introns. We also identified sequence motifs involved in hormone response and binding sites for RAV1 proteins and WRKY transcription factors through the prediction of cis-regulatory elements in the LhSorNPR1 promoter. Our gene expression analysis showed that LhSorNPR1 transcript levels significantly differed in various tissues, and that LhSorNPR1 expressions were induced by sodium salicylate, ethephon, and methyl jasmonate. Furthermore, we transformed LhSorNPR1 into Col-0 wild-type Arabidopsis to conduct function analysis, and we observed enhanced resistance to the bacterial pathogen Pseudomonas syringae pv. tomato DC3000 in the Arabidopsis expressing LhSorNPR1 gene. The enhanced disease resistance of LhSorNPR1 expressing plants could correlate to elevated expression levels in pathogenesis-related genes (PR1, PR2, and PR5) in vivo.
Keywords: LhSorNPR1, Lily, Systemic acquired resistance, Gene expression analysis, Transgenic Arabidopsis
Introduction
Signals from pathogen attacks at local infection sites are often transmitted to uninfected distal sites to protect plants from further invasion. This long-lasting, broad-spectrum defense response is referred to as systemic acquired resistance (SAR) (Fu and Dong 2013). The onset of SAR is accompanied by elevated levels of in vivo salicylic acid (SA) and induced expressions of pathogenesis-related (PR) genes (Gaffney et al. 1993; Rochon et al. 2006). Moreover, SA accumulation, at both infected and uninfected sites, has led to speculation that SA is the SAR signal molecule (Metraux et al. 1990; Rasmussen et al. 1991). Although much effort has been made in understanding the relationship between SA and SAR, there is to date no direct evidence of this. However, reducing SA levels by disrupting SA synthesis genes (Wildermuth et al. 2001) or by expressing bacterial salicylate hydroxylase genes (Vernooij et al. 1994) has compromised SAR phenotypes.
The non-expressor of PR genes 1 (NPR1) was first identified in Arabidopsis by screening mutants that were not sensitive to SA or its analogues (Cao et al. 1994, 1997). Analysis showed that the mutation of the NPR1 gene reduced SA-mediated PR genes expression and enhanced the susceptibility of plant species to pathogens (Delaney et al. 1995; Ryals et al. 1997). Furthermore, protein sequence analysis showed that NPR1 was characteristic of two domains, the BTB/POZ domain and the ankyrin repeat, both of which have been shown to mediate protein to protein interactions (Albagli et al. 1995; Aravind and Koonin 1999; Cao et al. 1997). However, DNA-binding domains (DBD), which regulate target gene expressions through promoter bindings, have not been detected in NPR1. The screening of interacting partners identified TGA transcription factors as being candidates that interplay with NPR1 (Kesarwani et al. 2007). The interaction between TGA family proteins and NPR1 revealed their roles in controlling expression of PR genes in SAR. NPR1 serves as a cofactor in enhancing the binding activity of TGA transcription factors in the promoter region to regulate PR genes expression (Despres et al. 2000).
As a key regulator of the plant defense signaling network (Pieterse and Van Loon 2004), NPR1 overexpression has been employed to enhance pathogen resistance in a number of plant species. For example, the ectopic expression of the StoNPR1 gene in potatoes provides enhanced resistance to the fungal pathogen Verticillium dahliae (Jue et al. 2014). In Vitis vinifera, VvNPR1.1 was identified as the functional ortholog of AtNPR1, and the overexpression of VvNPR1.1-GFP, a GFP labeled VvNPR1.1, in V. vinifera exhibited enhanced resistance to powdery mildew (Le Henanff et al. 2011). Moreover, transgenic carrots expressing AtNPR1 provided enhanced resistance to different types of pathogens (Wally et al. 2009), including necrotic fungi (Botrytis cinerea, Alternaria radicina, and Sclerotinia sclerotiorum) and biotrophic fungal pathogens (Erysiphe heraclei and Xanthomonas hortorum). Most previous studies have confirmed the critical role that NPR1 played in biotic stress, but only limited attention has been paid to the function of NPR1 on abiotic stress. When heterologously expressed in tobacco, AtNPR1 enhanced the oxidative stress tolerance of tobacco transgenic lines (Srinivasan et al. 2009). Similarly, increased tolerance to salt and osmotic stress was observed in tobacco plants overexpressing MhNPR1 (Zhang et al. 2014). These studies have uncovered the important roles that NPR1 play in both biotic and abiotic stresses, which will subsequently be used in future genetic engineering research.
Lilies are perennial bulbous plants that produce prized flowers. However, pathogens severely affect lily production in both bulbs and cut flowers. Lily bulbs can be infected by soil-borne pathogens, such as Fusarium oxyporum (Lecomte et al. 2016), Penicillium albocoremium, and Penicillium tulipae (Kim et al. 2006), causing scale, bulb base, and bud rot. Botrytis cinerea and Botrytis elliptica (Huang et al. 2012), causal agents of leaf and flower blight, severely affect cut flower quality and production. At the same time, viruses, such as the lily mottle virus (LMoV) and the lily symptomless virus (LSV), cause leaf mottling and stunted growth in lilies (Zhang et al. 2015a, b). Regardless of their differences in type, all these pathogens are major threats to lily production. Hence, a comprehensive understanding of the immunity-related mechanisms of lilies and their parasitic pathogens could facilitate lily bulb and flower production.
In this study, we characterized a NPR1-like gene from the oriental hybrid lily ‘Sorbonne’ (designated as LhSorNPR1). Sequence alignments showed that LhSorNPR1 shared domain characteristics of NPR1 orthologs, including the BTB/POZ domain and the ankyrin repeats. Analysis of the promoter sequence identified potential cis-regulatory elements (CREs) involved in the expression control of LhSorNPR1. Subsequent expression analysis showed that LhSorNRP1 was differentially expressed in various tissues, and its expression was responsive to sodium salicylate (SA), methyl jasmonate (MeJA), and ethephon (ETH). When consecutively expressed in Arabidopsis, LhSorNPR1 provided enhanced resistance to the bacterial pathogen Pseudomonas syringae pv. tomato DC3000 (Pst DC3000). Furthermore, the enhanced disease resistance of Arabidopsis transgenic lines could correlate to elevated expression levels for in vivo PR genes.
Materials and methods
Plant material and growth conditions
We used the oriental hybrid lily cultivar ‘Sorbonne’ in this study. We planted one bulb from 10 to 12 cm in diameter per pot (pot = 25 cm in diameter) containing peat moss as a growing substrate. We stored all bulbs used in this study at 4 °C for over 2 months to end the lily dormant state before the experiment started. We then transferred all pots to a growth chamber that was maintained at optimum growth conditions (16 h light/8 h dark; 22 °C).
Genomic DNA extraction, total RNA isolation, and first-strand cDNA synthesis
We conducted genomic DNA extraction as previously described (Clarke 2009) with slight modifications. In brief, we milled 500 mg of leaf tissue to a powder using a mortar and pestle. The powdered tissue was then homogenized in an extraction buffer (2% CTAB, 1% PVP, 1.4 M NaCl, 10 mM EDTA, 100 mM Tris–HCl, 1% β-mercaptoethanol) and incubated for 2 h at 65 °C. After incubation, we added 1 volume of chloroform to the mixture and centrifuged it to remove proteins. Residual RNA that could interfere with downstream PCR was degraded using RNase A. The quality of the precipitated DNA was verified using 1% (w/v) agarose gel electrophoresis and a Nanodrop 2000c UV–Vis spectrophotometer (Thermo Fisher Scientific, USA).
We conducted total RNA isolation using the RNAprep Pure Kit (for plants) (TIANGEN Corporation, Beijing, China) following the manufacturer’s instructions. We removed residual DNA in the isolated RNA using the DNase I treatment. We determined concentrations of extracted RNA using Nanodrop 2000c, and we assessed RNA quality applying 260/280 and 260/230 absorbance ratios. Furthermore, we reversely transcribed 5 µg of total RNA into cDNA using the PrimeScript II First Strand cDNA Synthesis Kit (Takara Biotechnology, Dalian, China). For cDNA synthesis of rapid amplification of cDNA ends (RACE) cloning, we used 1 µg of total RNA for first strand cDNA synthesis. RACE cloning was conducted with the SMARTer® RACE 5′/3′ Kit (Clontech Laboratories, USA) according to the manufacturer’s instructions.
Full-length LhSorNPR1 cloning
To obtain the partial coding sequence of LhSorNPR1, we designed a pair of degenerate primers (NPR1-deg-F and NPR1-deg-R; Table 1) based on the conserved regions of NPR1 orthologs. We designed gene-specific primers (GSP) for RACE (Table 1) based on the partial LhSorNPR1 coding sequence obtained. We conducted primary RACE PCR in a 25 µL reaction mixture containing 20 ng of 3′ RACE template cDNA (or 5′ RACE template cDNA), 1.0 µmol L−1 of 3′ RACE-GSP-1 (or 5′ RACE-GSP-1), 200 µmol L−1 of dNTPs, 0.04 µmol L−1 Long Universal Primer A (Long UP), 0.2 µmol L−1 Short Universal Primer A (Short UP), and 1.5 U Taq DNA polymerase. We diluted PCR products of primary PCR by a ratio of 50:1 and used this as a template for the next step (Nested PCR). We conducted Nested PCR for 5′ RACE (or 3′ RACE) in a 25 µL reaction mixture containing 2 µL of diluted primary 5′ RACE (or 3′ RACE) PCR products, 1.0 µmol L−1 of 5′ RACE-GSP-2 (or 3′ RACE-GSP-2), 1.0 µmol L−1 of Nested Universal Primer A, 200 µmol L−1 of dNTPs, and 1.25 U Taq DNA polymerase. Amplification conditions for primary and nested RACE PCR followed the same protocol: 94 °C for 30 s, 68 °C for 30 s, and 72 °C for 3 min for 25 cycles. PCR products were gel purified and ligated into pMD18T vectors (Takara Biotechnology, Dalian, China) for sequencing (Sangon Biotech, Shanghai, China). We obtained the full-length sequence of LhSorNPR1 by assembling the 5′ RACE sequence, the partial coding sequence, and the 3′ RACE sequence. We used the primer set (LhSorNPR1-full-length-F and LhSorNPR1-full-length-R) for full-length cDNA and full-length genomic DNA LhSorNPR1 cloning.
Table 2.
NPR1-like protein sequences retrieved from GenBank used for phylogram construction
| Indentifier | Accession number | Species |
|---|---|---|
| AtNPR1 | NP_176610 | Arabidopsis thaliana |
| AtNPR2 | NP_194342 | A. thaliana |
| AtNPR3 | NP_199324 | A. thaliana |
| AtNPR4 | NP_193701 | A. thaliana |
| OsNPR1 | AAX18700 | Oryza sativa |
| OsNPR2 | ABE11616 | O. sativa |
| OsNPR3 | ABE11618 | O. sativa |
| OsNPR5 | ABE11622 | O. sativa |
| GmNPR1-1 | ACJ45013 | Glycine max |
| GmNPR1-2 | ACJ45015 | G. max |
| MtNPR1 | XP_003594464 | Medicago truncatula |
| PdNPR1-1 | AEY99652 | Populus deltoides |
| PdNPR2 | AEE81755 | P. deltoides |
| PpNPR1-1 | ABK62792 | Pyrus pyrifolia |
| PtNPR1.1 | XP_002308281 | Populus trichocarpa |
| PtNPR1-like | XP_002323261 | P. trichocarpa |
| MhNPR1 | ACU78081 | Malus hupehensis |
| HaNPR1 | AAT57642] | Helianthus annuus |
| MdNPR1 | ACJ04030 | Musa spp. ABB |
| SbNPR1 | XP_002455011 | Sorghum bicolor |
| HvNPR1 | CAJ19095 | Hordeum vulgare subsp. vulgare |
| PaNPR1 | AEP68016 | Phalaenopsis aphrodite subsp. formosana |
| BvNPR1 | AAT57640 | Beta vulgaris |
| VvNPR1.1 | XP_002281475 | Vitis vinifera |
| IbNPR1 | ABM64782 | Ipomoea batatas |
| NtNPR1 | AAM62410 | Nicotiana tabacum |
| CaNPR1 | ABG38308 | Capsicum annum |
| LeNPR1 | AAT57637 | Solanum lycopersicum |
| PhNPR-like1 | XP_001757508 | Physcomitrella patens |
| PhNPR-like2 | XP_001759240 | P. patens |
| MpNPR1-1 | ACC77697 | Malus x domestica |
Table 1.
List of primers used in the experiment
| Experiment | Primer name | Primer sequence (5′–3′) |
|---|---|---|
| NPR1 degenerate PCR | NPR1-deg-F | CAYCGNGCNCTNGAYTCNGAYGA |
| NPR1-deg-R | CGNCGNCCNAGYTCNACNGT | |
| LhSorNPR1 5′ RACE | LhSorNPR1-5RACE-GSP-1 | CAGTCGGGAGCAGCTCTACAAGCGC |
| LhSorNPR1-5RACE-GSP-2 | CGCTGAAACAGCGAGACCAGCTCGG | |
| LhSorNPR1 3′ RACE | LhSorNPR1-3RACE-GSP-1 | GGGTGACGAGAATCCACCGTGCTCTGG |
| LhSorNPR1-3RACE-GSP-2 | GGAGCGGGAGATGATGAGGAACCCC | |
| LhSorNPR1 full-length cloning | LhSorNPR1-full-length-F | ATGGCCGACGCCGCCGAG |
| LhSorNPR1-full-length-R | TCATTCTTCCATATCTACCAGACAGAC | |
| 5′ RACE and 3′ RACE | Universal Primer A (Long) | CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT |
| Universal Primer A (Short) | CTAATACGACTCACTATAGGGC | |
| 5′ RACE and 3′ RACE | Nested Universal Primer A | AAGCAGTGGTATCAACGCAGAGT |
| LhSorNPR1 promoter cloning | AC1 | ACGATGGACTCCAGAG |
| LAD1-1 | ACGATGGACTCCAGAGCGGCCGCVNVNNNGGAA | |
| LAD1-2 | ACGATGGACTCCAGAGCGGCCGCBNBNNNGGTT | |
| LAD1-3 | ACGATGGACTCCAGAGCGGCCGCVVNVNNNCCAA | |
| LAD1-4 | ACGATGGACTCCAGAGCGGCCGCBDNBNNNCGGT | |
| LhSorNPR1-promoter-TAIL-1 | GGCGCGTGAGGCTGACGACCTCGAGG | |
| LhSorNPR1-promoter-TAIL-2 | ACGATGGACTCCAGTCCGGCCTGGTGCCGTTGGAGACGTAGGATGAGGT | |
| LhSorNPR1-promoter-TAIL-3 | GGACCACTCGGCGGCGTCGGCCAT | |
| LhSorNPR1 qPCR | LhSorNPR1 qPCR-F | CTTGATAAGTTCTTGGAGGACGAT |
| LhSorNPR1 qPCR-R | GATGTAGACGATGATGACGATGAT | |
| Construction of the LhSorNPR1 expression vector | LhSorNPR1-OE-F | TAGGATCCATGGCCGACGCCGCCGA |
| LhSorNPR1-OE-R | CGGGTACCTCATTCTTCCATATCTACCAGACAG | |
| AtPR1 qPCR | AtPR1-qPCR-F | GCTCTTGTTCTTCCCTCGAAAG |
| AtPR1- qPCR-R | GCCTCTTAGTTGTTCTGCGTAGCT | |
| AtPR2 qPCR | AtPR2- qPCR-F | ATCTCCCTTGCTCGTGAATCTCT |
| AtPR2- qPCR-R | TCGAGATTTGCGTCGAATAGG | |
| AtPR5 qPCR | AtPR5- qPCR-F | GAGGATCGGGAGATTGCAAA |
| AtPR5- qPCR-R | CTCCACGGCAGCAATATTGA | |
| AtACTIN2 qPCR | AtACTIN2-F | ACGGTAACATTGTGCTCAGTGGTG |
| AtACTIN2-R | CTTGGAGATCCACATCTGCTGGA |
Restriction sites for vector construction are underlined
LhSorNPR1 promoter isolation and cis-regulatory elements prediction
We conducted promoter cloning using hiTAIL-PCR according to the protocol previously proposed (Liu and Chen 2007). Initially, we conducted preamplifications in 25 µL reaction mixtures, each containing 200 µmol L−1 of dNTPs, 0.3 µM LhSorNPR1-promoter-TAIL-1, 1.0 µM of any one of the LAD primers (Table 1), 1.25 U Taq DNA polymerase, and 50 ng of the lily genomic DNA. We diluted PCR preamplification products by a ratio of 40:1 and used it as a template for the next step (primary amplification). We conducted PCR reactions for primary amplification in a 25 µL reaction mixture containing 200 µmol L−1 of dNTPs, 0.3 µM LhSorNPR1-promoter-TAIL-2 and AC1, 1 µL of diluted preamplification product, and 1.25 U Taq DNA polymerase. We conducted secondary amplifications of TAIL PCR in a 25 µL reaction volume, each containing 200 µmol L−1 of dNTPs, 0.3 µM LhSorNPR1-promoter-TAIL-3 and AC1, 1 µL of primary product (diluted by a ratio of 10:1), and 1.25 U Taq DNA polymerase. Thermal conditions for all hiTAIL-PCR reactions followed the protocol proposed by Liu and Chen. We submitted the sequence of the isolated LhSorNPR1 promoter to PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) and PLACE (http://www.dna.affrc.go.jp/PLACE/) databases for CREs prediction.
In silico analysis of LhSorNPR1
We predicted the open reading frame (ORF) of LhSorNPR1 using the ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/). Homology of the LhSorNPR1 putative protein sequence to other proteins was confirmed by querying the National Center for Biotechnology Information (NCBI) database using the Basic Local Alignment Search Tool (BLAST) (http://blast.ncbi.nlm.nih.gov/Blast.cgi). We calculated the theoretical isoelectric point (pI) and molecular weight (MW) of LhSorNPR1 using the Compute pI/Mw tool (http://web.expasy.org/compute_pi/). The exon/intron organization of NPR1 genes was generated using the Gene Structure Display Server 2.0 (http://gsds.cbi.pku.edu.cn/). We conducted LhSorNPR1 sequence alignment with other NPR1 orthologs using the ClustalW program (http://www.ebi.ac.uk/clustalw). We constructed the phylogram of LhSorNPR1 and other NPR1 homologues from different species applying the neighbor-joining method using Mega 4.1 software (Tamura et al. 2007).
Expression analysis of LhSorNPR1 in different tissues and under various phytohormone treatments
For the tissue-specific expression of LhSorNPR1, we sampled roots, stems, leaves, petals, and scales from 2-month-old lilies that had fully bloomed. For gene expression analysis of LhSorNPR1 under different phytohormone treatments, we subjected 30-day-old seedlings of lily plants to foliar application of 5 mM ETH, 10 mM SA, or 20 mM MeJA. For the latter treatment, MeJA was first dissolved in ethanol and then diluted to the desired concentration in ddH2O. Pure water containing the same concentration of ethanol (1%) was used as the mock for MeJA treatment. For ETH and SA treatments, we used a foliar spray of pure water as the mock treatment. We sampled leaves 0, 2, 4, 8, 12, and 24 h after the phytohormone (or mock) treatments and subjected them to total RNA extraction. Furthermore, we reversely transcribed 500 ng of total RNA into cDNA using the HiScript II Q RT SuperMix for qPCR kit (Vazyme Biotech Co., Nanjing, China), following the manufacturer’s instructions. We diluted the synthesized cDNA with RNase-free water and used it as a template for the subsequent qPCR test. We conducted real-time PCRs for gene expression level quantification in a reaction of 20 µL using the AceQ qPCR SYBR Green Master Mix kit (Vazyme Biotech Co., Nanjing, China). We designed the LhSorNPR1 primer set for qPCR (LhSorNPR1 qPCR-F and LhSorNPR1 qPCR-R) using OligoArchitect (http://www.sigmaaldrich.com/china-mainland/zh/technical-documents/articles/biology/oligoarchitect-online). We used the polyubquitin4 gene (GenBank accession no. DW718023) of the oriental hybrid lily ‘Sorbonne’ as the internal standard. We conducted qPCR reactions using the MX3000P qPCR thermo cycler (Stratagene, USA). We used the following thermal conditions for amplification: pre-denaturation for 10 min at 95 °C, followed by 40 cycles at 94 °C for 15 s and 60 °C for 30 s. After amplification, we charted melting curves (from 60 to 95 °C) to detect the existence of non-specific amplicons. We determined application efficiency from the slope of the plotted LhSorNPR1 standard curve. We conducted calculations of gene expression levels using the relative expression software tool (REST) 2009, and we used the P(H1) test for statistical analysis of gene expressions (Pfaffl et al. 2002).
Overexpression of LhSorNPR1 in Arabidopsis
To construct the LhSorNPR1-pBI121 overexpression vector, ORF primers (Table 1) with BamHI and KpnI sites were designed, and the amplified PCR product was inserted into a modified pBI121 vector preserved in our lab. After verification through sequencing, we introduced the LhSorNPR1-pBI121 recombinant plasmid into the Agrobacterium tumefaciens strain GV3101 using the heat-shock method. We used the resulting A. tumefaciens strain containing the LhSorNPR1-pBI121 vector to transform Arabidopsis Col-0 accession plants as previously described (Clough and Bent 1998). We screened Arabidopsis transformants on an Murashige and Skoog (MS) medium containing 50 µg/ml kanamycin. We used T3 homozygous lines for the subsequent experiments.
Expression analysis of pathogenesis-related genes in transgenic Arabidopsis plants
We extracted total RNA of 3-week-old Arabidopsis seedlings and reversely transcribed them into cDNA as described above. We quantified expression levels of PR genes (PR1, PR2, and PR5) by qPCR using the Arabidopsis actin gene (AtACTIN2) as the internal standard. We conducted calculations of gene expression levels and statistical analysis using REST 2009 software as described above.
Challenges of transgenic Arabidopsis to Pseudomonas syringae pv. tomato DC3000
The Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) bacteria was streaked from a glycerol stock on agar plates (low salt LB) supplemented with 50 µg mL−1 rifampicin. After 2 d incubation at 28 °C, a single colony was inoculated in the liquid LB supplemented by 50 µg mL−1 rifampicin and incubated on a shaker, set at 28 °C for 8 to 12 h until OD600 reached 0.6 (Katagiri et al. 2002). The culture was centrifuged at 5000 g for 10 min to remove the supernatant, and the bacteria pellet was then resuspended in sterile water. We adjusted the bacteria concentration in a water suspension to ~106 cfu mL−1 (OD600 = 0.001), and this was used for leaf infiltration of 3-week-old plants (applying a needleless syringe). In order to further compare bacterial infection quantitatively, discs were punched from the leaves of wild-type and transgenic plants 3 days after infiltration. The leaf punches were surface sterilized and then milled in 1 mL distilled water using a mortar and pestle. The resultant suspensions were serially diluted and plated on LB agar plates supplemented in 50 µg mL−1 rifampicin. After being incubated at 28 °C for 2 days, the number of colonies was counted to calculate the colony forming units (CFU) per square centimeter of leaf tissue. Five days after inoculation, the leaves of Col-0 wild-type and LhSorNPR1 transgenic plants were photographed to compare phenotypic changes.
Results
Isolation of the full-length LhSorNPR1 cDNA
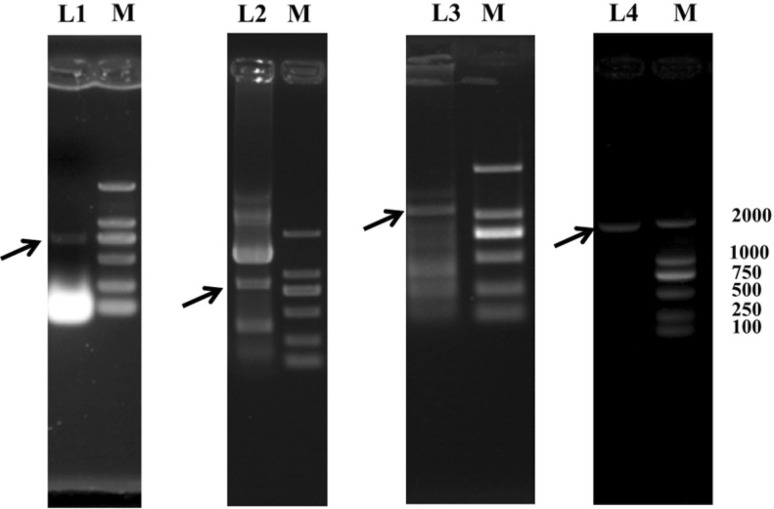
To obtain the partial coding sequence of the NPR1 gene in lilies, we designed a pair of degenerate primers (Table 1) based on the conserved regions of the NPR1 orthologs from different plants. As predicted, we amplified a DNA fragment ~700 bp in the degenerate PCR (Fig. 1; L1). Using the partial coding sequence obtained, we designed 5′ RACE and 3′ RACE GSP primers. As indicated by the arrows in Fig. 1, we obtained amplicons of 864 bp and 1143 bp in size for the 5′ RACE (Fig. 1; L2) and the 3′ RACE (Fig. 1; L3) PCRs, respectively. We verified the assembled full-length LhSorNPR1 sequence using PCR, and we amplified a 1854 bp DNA fragment corresponding to the LhSorNPR1 ORF (Fig. 1; L4). The ORF encoded a protein of 617 amino acids with a predicted pI of 5.95 and a theoretical molecular mass of 68.87 kDa. The 5′ and 3′ untranslated regions (UTR) flanking the ORF were 140 and 250 bp, respectively. The verified full-length LhSorNPR1 sequence was submitted to GenBank and deposited under the accession number KY073343.
Fig. 1.
Cloning of the LhSorNPR1 gene by RACE. M: DL2000 marker; L1: product of degenerate PCR. The amplified partial coding sequence of LhSorNPR1 is indicated by an arrow. L2: 5′ LhSorNPR1 RACE products. The 5′ end fragment amplified by Nested PCR is indicated by an arrow. L3: 3′ LhSorNPR1 RACE products. The 3′ end fragment amplified by Nested PCR is indicated by an arrow. L4: the 1854 bp LhSorNPR1 full-length cDNA amplified. The fragment amplified is indicated by an arrow
Genomic structure of LhSorNPR1
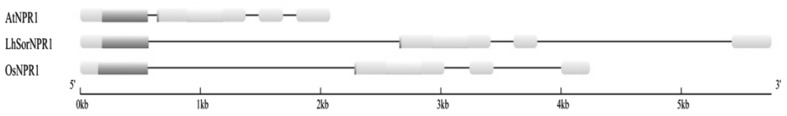
Using the LhSorNPR1 full-length cloning primers (Table 1), we successfully amplified the genomic DNA fragment of LhSorNPR1, which was 5751 bp in length from the start codon to the stop codon. Further sequence alignments of LhSorNPR1 cDNA and genomic DNA showed that there were four exons in the LhSorNPR1 genomic DNA fragment, which essentially means that these exons were flanked by three introns. The exon/intron organization of NPR1 genes was similar in different plants species, since NPR1 orthologs in both Arabidopsis and rice share the same genomic structure with LhSorNPR1 (Fig. 2). The conservation in genomic DNA structure suggested that LhSorNPR1 could provide the same function as their orthologs.
Fig. 2.
Comparison of the genomic structure of LhSorNPR1 between AtNPR1 (U87794) and OsNPR1 (DQ450948). Exons (boxes) are connected by introns (black lines) between them. The BTB/POZ domain and ankyrin repeats domain are represented by boxes shaded in purple and green respectively (color figure online)
Protein sequence analysis of LhSorNPR1
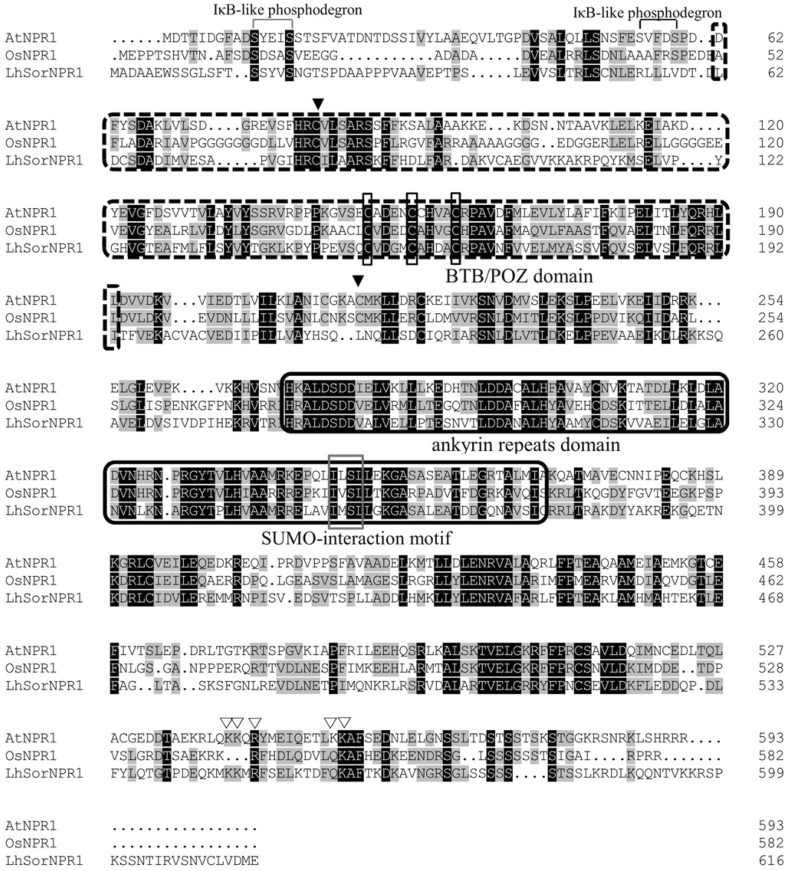
Multiple alignments of the deduced LhSorNPR1 protein sequence with NPR1 orthologs in Arabidopsis and rice showed that the NPR1 protein family shared several domains, such as the BTB/POZ domain (Fig. 3; dashed-line box) and the ankyrin repeats (Fig. 3; solid-line box). The BTB/POZ domain functions in the homodimerization of AtNPR1 in Arabidopsis, and the ankyrin repeats mediate interactions with the TGA2 transcription factor to control PR genes expression. In addition to these two domains that are well conserved, five cys residues (C82, C150, C155, C160, and C216) have been shown to play important roles in the oligomerization of AtNPR1 (Mou et al. 2003; Tada et al. 2008). Among these five residues, C82 and C216 were critical in the oligomerization of AtNPR1 (Fig. 3; solid triangles). Moreover, C82, C150, C155, and C160 were conserved in LhSorNPR1 (as shown in Fig. 3), but gln was substituted for C216. Post-translational modifications (PTM), such as phosphorylation, ubiquitination and sumoylation, also have profound impact on plant immunity regulated by NPR1. The S11/S15 (in AtNPR1) IκB-like phosphodegron (Spoel et al. 2009), whose phosphorylation was required for AtNPR1 ubiquitination and proteasome-mediated degradation was conserved in LhSorNPR1 (S15/S19; Fig. 3). While the second IκB-like motif (S55/S59 in AtNPR1), whose phosphorylation was critical for the inhibition of AtNPR1 sumoylation and degradation (Saleh et al. 2015), was not conserved in LhSorNPR1(Fig. 3). However, the small ubiquitin-like modifier (SUMO) interaction motif (SIM), which mediated conjugation of SUMO3 to the lysine residue(s) in AtNPR1, was conserved in LhSorNPR1. The translocation of NPR1 required the participation of the nuclear localization signal (NLS) at the C-terminus. Five basic residues (Fig. 3; hollow triangles), which were found to be critical for the nuclear localization of AtNPR1, were conserved in LhSorNPR1. The conservation of these functional domains in the NPR1 protein family implies that LhSorNPR1 could render similar functions for NPR1 orthologs in lilies.
Fig. 3.
LhSorNPR1 sequence alignment with NPR1s in Arabidopsis and rice. LhSorNPR1 was aligned with AtNPR1 (Arabidopsis thaliana NPR1; NP176610) and OsNPR1 (Oryza sativa NPR1; ABE11614) using ClustalW. Identical residues are shaded in black, and highly similar residues are shaded in gray. Dashed-line boxes represent the BTB/POZ domain. Solid-lines represent the ankyrin repeats. Solid triangles represent C82 and C216, which are essential to AtNPR1 oligomerization in cytoplasm. Rectangular boxes represent C150, C155, and C160, which also function in oligomer-monomer exchanges. Hollow triangles represent amino acid residues required for the nuclear translocation of AtNPR1. The small ubiquitin-like modifier (SUMO) interaction motif (SIM), which mediate conjugation of SUMO3 to the lysine residue(s) in AtNPR1, is highlighted by a red box. The S11/S15 IκB-like phosphodegron, whose phosphorylation is required for AtNPR1 ubiquitination and degradation, is highlighted with a blue bracket. The second IκB-like motif, S55/S59, whose phosphorylation is critical for the inhibition of AtNPR1 sumoylation and degradation, is highlighted with a black bracket (color figure online)
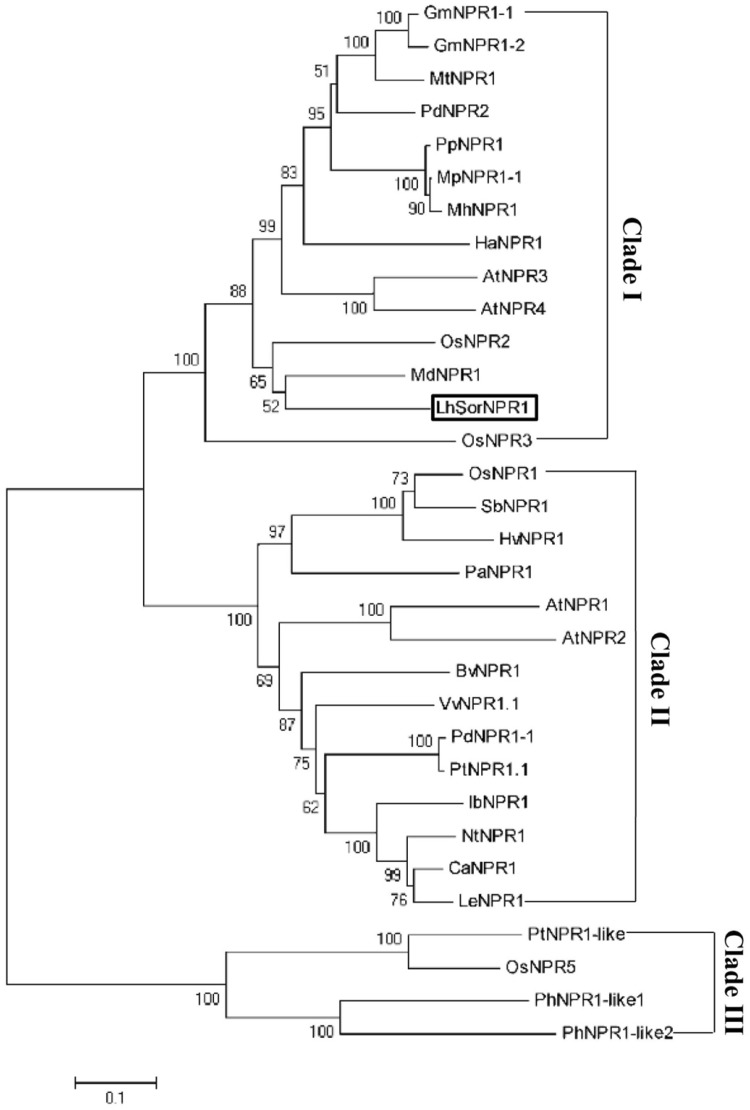
Phylogenetic analysis
Protein sequences of 32 NPR1 homologues in different plants were retrieved from Genbank to construct the phylogram. As shown in Fig. 4, NPR1-like genes from these species mainly grouped into three clades. LhSoNPR1 was most closely related to MdNPR1, a NPR1 ortholog originated from Musa spp. ABB.
Fig. 4.
Phylogram of LhSorNPR1 and other NPR1 homologues from different species. The Phylogenetic tree was constructed with MEGA4 using neighbor-joining (NJ) method. Branch length is proportional to time of divergence. The scale bar represents a 5% change. The accession numbers corresponding to all protein sequences used in the phylogram were summaried in Table 2
Analysis of potential cis-regulatory elements involved in the regulation of gene expression for the LhSorNPR1 promoter
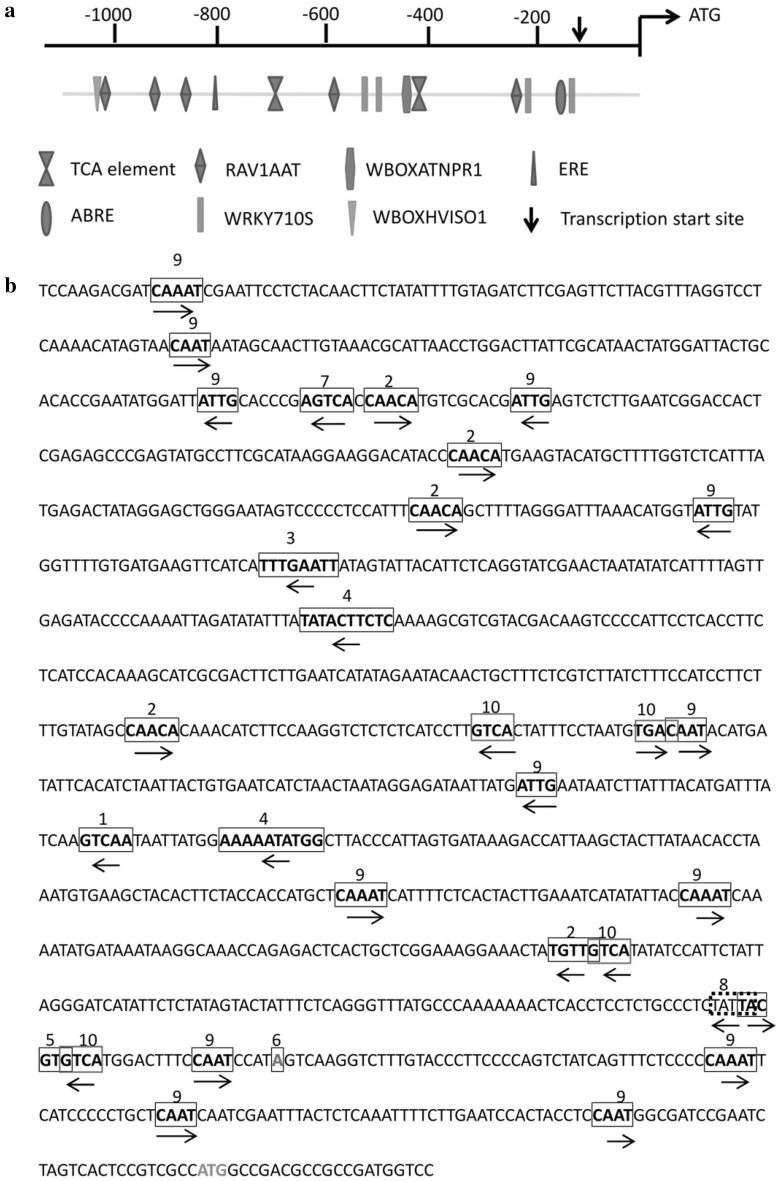
The LhSorNPR1 promoter sequence, which corresponds to a DNA fragment of 1196 bp upstream of the start codon, was successfully isolated by employing the hiTAIL-PCR method. Subsequent sequence alignments of the promoter and the LhSorNPR1 5′ UTR region showed that the transcription start site (TSS) of LhSorNPR1 was located at −140 bp upstream of the start codon (Fig. 5b). We found a TATA box (Fig. 5b), which is the binding site of general transcription factors or histones, located at −29 bp upstream of the TSS (an adenine). Several CAAT boxes, as shown in Fig. 5b, were also found in the LhSorNPR1 promoter. In addition to these two CREs involved in general transcription, the presence of hormone responsive elements indicated that the LhSorNPR1 expression was subjected to regulation by phytohormones in different ways (Fig. 5a). These CREs include the TCA element involved in SA response, the ABRE element involved in ABA response, and the ERE element involved in ethylene response. We also found several CREs types involved in defense signaling in the LhSorNPR1 promoter. For example, we detected five potential RAV1AAT element sites (Fig. 5a) that could enable the binding of RAV1 proteins. RAV1 transcription factors regulated expressions of multiple defense-related genes through the binding of RAV1AAT CREs in the promoter region (Sohn et al. 2006). The presence of W boxes in the LhSorNPR1 promoter showed that WRKY family proteins were involved in NPR1 expression regulation in lilies. Furthermore, we detected a WBOXATNPR1 motif, the W box for WRKY protein binding in AtNPR1, in the LhSorNPR1 promoter. This seemed to imply that similar mechanisms should exist in LhSorNPR1 in relation to the WRKY transcription factor contributed expression control. In addition to the WBOXATNPR1 motif, we also detected W boxes in two other different types (WRKY71OS and WBOXHVISO1) in the LhSorNPR1 promoter. The sequence of LhSorNPR1 promoter and all potential cis-elements predicted are shown in Fig. 5b.
Fig. 5.
Prediction of Cis-regulatory elements in LhSorNPR1 promoter. a Schematic diagram of cis-regulatory elements involved in the regulation of LhSorNPR1 expression in the promoter region. Distribution of core elements involved in LhSorNPR1 regulation are shown as indicated. The scale on top of the bar represents the distance from the LhSorNPR1 start codon. b Sequence analysis of LhSorNPR1 promoter. 1: WBOXATNPR1, 2: RAV1AAT, 3: ERE, 4: TCA-element, 5: ABRE, 6: Transcription start site, 7: WBOXHVISO1, 8: TATA BOX, 9: CAAT BOX, 10: WRKY71OS. We obtained a 1196 bp DNA fragment upstream of the start codon using hiTAIL-PCR. Cis-regulatory elements in the obtained sequence were predicted by querying PlantCARE and PLACE databases
Expression analysis of LhSorNPR1 in different tissues and under various phytohormone treatments
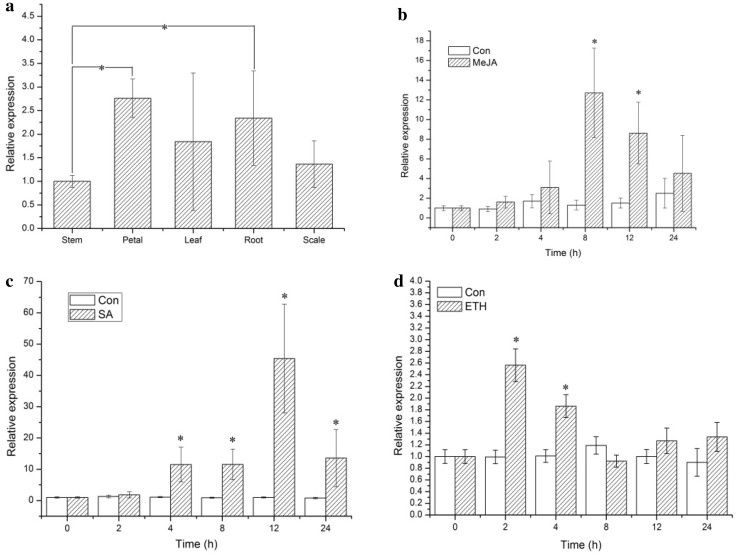
For LhSorNPR1 expression analysis (using qPCR), we first plotted standard curves and dissociation curves to calculate the amplification efficiency and to test the amplification specificity. We calculated LhSorNPR1 gene expression levels in different tissues relative to the calibrator (the stem expression level was set at 1.0). As shown in Fig. 6a, we found that LhSorNPR1 expression levels in petals and roots were significantly higher than in stems (by a magnitude of 2.76 and 2.33, respectively). However, LhSorNPR1 transcript levels in leaves and scales were not significantly different from stems.
Fig. 6.
Expression analysis of LhSorNPR1 in different tissues and under various phytohormone treatments. a The tissue-specific expression of LhSorNPR1; b the temporal expression of LhSorNPR1 in response to the methyl jasmonate (MeJA) treatment; c the temporal expression of LhSorNPR1 in response to the sodium salicylate (SA) treatment; d the temporal expression of LhSorNPR1 in response to the ethephon (ETH) treatment. We used the LhSorNPR1 expression at 0 h as a calibrator (designated as 1.0) to determine the relative expression of the target gene at different time points. Values are presented as mean ± standard deviation (SD) for the three replicates. Bars labeled with an asterisk (*) indicate significant differences from the control at P < 0.05 (the REST statistical randomization test)
Phytohormones, such as JA, SA, and ethylene, are major contributors to biotic stress signaling in plants. Hence, we investigated temporal expression patterns of LhSorNPR1 in response to these three phytohormones. Exogenous application of MeJA, a functional analogue of JA, stimulated LhSorNPR1 transcript accumulation in leaves. As shown in Fig. 6b, the LhSorNPR1 expression significantly increased with a lapse in MeJA treatment time, and reached its peak level 8 h after treatment began (at a magnitude of 12.7 compared to the control). For the SA treatment (Fig. 6c), the LhSorNPR1 expression was significantly up-regulated 4 h after treatment began, reaching a magnitude of 11.49 compared to the control, and then attained its highest expression level at 12 h (at a magnitude of 45.4 compared to the control). The exogenous application of ETH stimulated the LhSorNPR1 expression, producing with a relatively lower intensity compared to MeJA and SA. After 2 h ETH treatment, the LhSorNPR1 expression significantly increased, reaching a magnitude of 2.56 compared to the control. Although the LhSorNPR1 expression was also significantly different from the control after 4 h, we observed no significant time point changes in gene expression levels after this point in time. These findings suggested that multiple phytohormones could induce the LhSorNPR1 expression, and this inducibility could be correlated to hormone-responsive CREs in the promoter.
Enhanced resistance to Pseudomonas syringae pv. tomato DC3000 was observed in Arabidopsis expressing LhSorNPR1
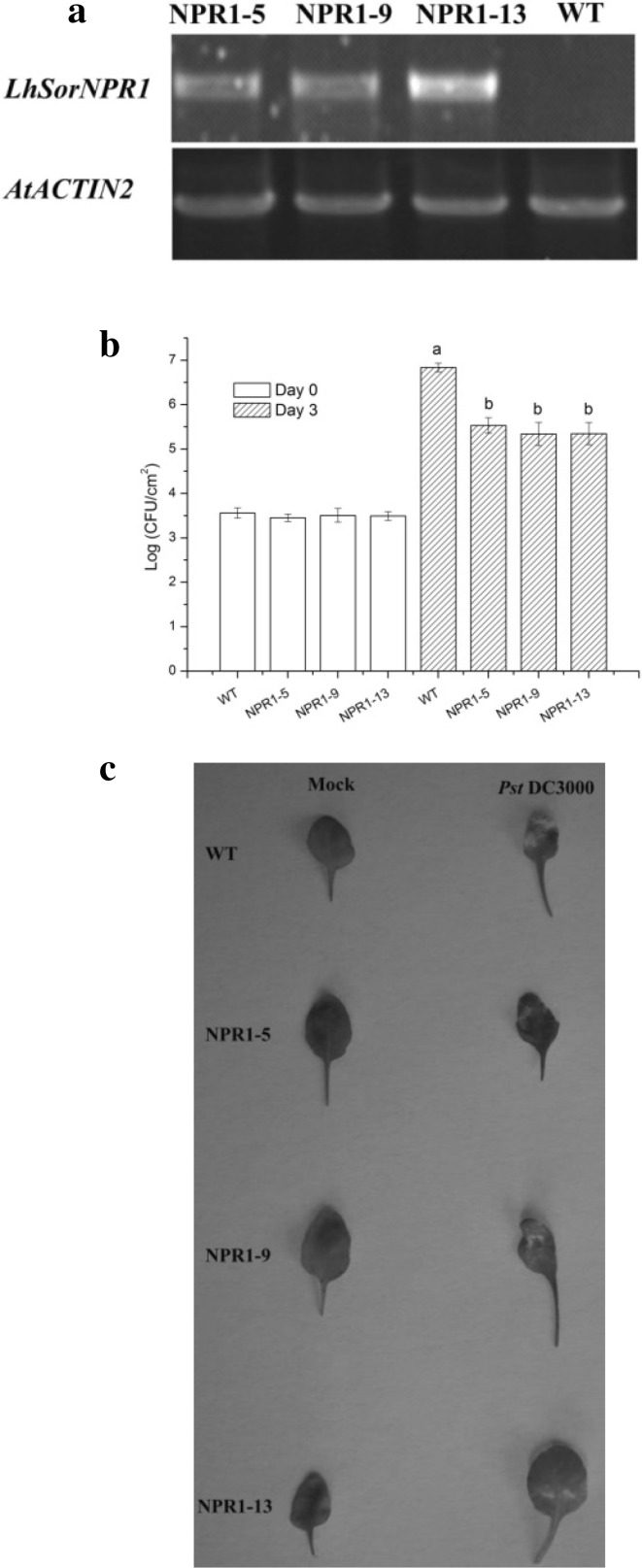
Transgenic Arabidopsis lines constitutively expressing LhSorNPR1 were produced to validate the gene function. We transformed LhSorNPR1 into the Col-0 wild-type Arabidopsis under the control of CaMV 35S promoter. As shown in Fig. 7a, we observed high LhSorNPR1 expressions in three transgenic lines of LhSorNPR1, but the LhSorNPR1 expression was not detectable in the untransformed Col-0 wild-type plant. We then challenged these three transgenic lines (NPR1-5, NPR1-9, and NPR1-13) with virulent Pst DC3000. Three days after inoculation, the number of Pst DC3000 per cm2 in leaf discs was quantified to compare pathogen multiplication statistically. As shown in Fig. 7b, the Col-0 wild-type and LhSorNPR1 transgenic lines did not differ in the number of Pst DC3000 that were quantified immediately after infiltration (0 day). However, Pst DC3000 in the Col-0 wild-type multiplied to a significantly higher level than observed for LhSorNPR1 transgenic lines 3 days after inoculation (3 days). Five days after inoculation, Col-0 wild-type Arabidopsis leaves showed severe signs of necrosis (Fig. 7c). The three LhSorNPR1 transgenic lines, however, displayed less severe signs of necrotic phenotypes upon Pst DC3000 infection. The enhanced disease resistance provided by the LhSorNPR1 overexpression in Arabidopsis suggested that LhSorNPR1 had a NPR1 ortholog encoded within it that promoted lily disease resistance.
Fig. 7.
Resistance of Arabidopsis LhSorNPR1 transgenic lines to Pseudomonas syringae pv. tomato DC3000. a Detection of the LhSorNPR1 expression in transgenic Arabidopsis; total RNA was isolated from three T3 homozygous lines and the Col-0 wild-type plant; after reverse transcription, the full-length LhSorNPR1 sequence was specifically amplified from LhSorNPR1 transgenic plant species, but we did not detect LhSorNPR1 expressions from the Col-0 wild-type. b Multiplication of Pst DC3000 in leaves of LhSorNPR1 transgenic lines and the Col-0 wild-type plant; we collected leaf punches at infection sites 0 and 3 days after inoculation to quantify bacterial growth. Values represent mean ± standard deviation (SD) for the three replicates. Bars labeled with different letters differed significantly (P < 0.05; t test). c Disease symptoms of three independent transgenic lines and the Col-0 wild-type plant; leaves of transgenic lines and the wild-type plant species were inoculated with Pseudomonas syringae pv. tomato DC3000 (OD600 = 0.001) or sterile water (mock); we observed disease symptoms 5 days after inoculation
LhSorNPR1 overexpression elevated the transcript levels of pathogenesis-related genes in Arabidopsis
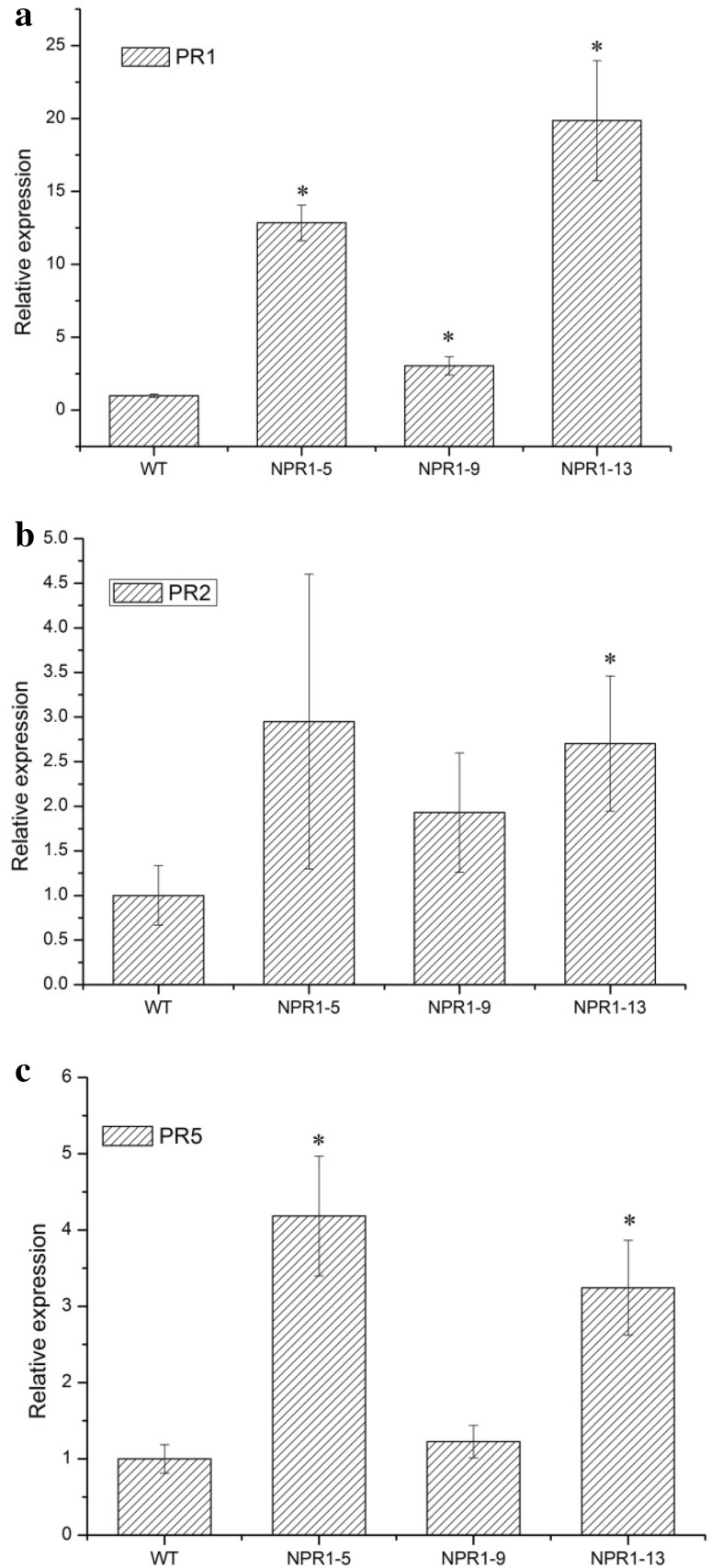
To determine whether the LhSorNPR1 overexpression elevated expression levels of PR genes, we quantified transcript levels of three marker genes (PR1, PR2, and PR5) for SA-mediated defense pathways (using qPCR). As shown in Fig. 8a, we found that LhSorNPR1 expression significantly enhanced the PR1 expression in all three transgenic lines. The transcript level of PR2 was significantly elevated in NPR1-13 transgenic line, reaching a magnitude of 2.7 compared to the wild-type control (Fig. 8b). For the PR5 expression, the NPR1-5 and NPR1-13 transgenic lines showed a significant increase in PR5 transcript level compared to the WT control (Fig. 8c). Elevated transcript levels of PR1, PR2, and PR5 in conjunction with the enhanced resistance to Pst DC3000 in LhSorNPR1 transgenic lines indicated that LhSorNPR1 disease resistance could be correlated to the enhanced expression of PR genes.
Fig. 8.
Expression analysis of PR genes in Arabidopsis transgenic lines. a PR1 transcript levels in LhSorNPR1-expressing transgenic plants and untransformed Col-0 wild-type Arabidopsis. b PR2 transcript levels in LhSorNPR1-expressing transgenic plants and untransformed Col-0 wild-type Arabidopsis. c PR5 transcript levels in LhSorNPR1-expressing transgenic plants and untransformed Col-0 wild-type Arabidopsis. We used the expression level in the Col-0 wild-type Arabidopsis as a calibrator (set at 1.0) to calculate the relative expression of PR1, PR2, and PR5 in transgenic lines. Values are shown as mean ± standard deviation (SD) for the three replicates. Bars labeled with an asterisk (*) indicate significant differences from the Col-0 wild-type control at P < 0.05 (REST statistical randomization test)
Discussion
The aim of this study was to characterize LhSorNPR1, a novel member of the NPR1 protein family from the oriental hybrid lily ‘Sorbonne’. Multiple sequence alignment showed that LhSorNPR1 exhibited a high level of identity and similarity to other NPR1 orthologs in plant species. For example, LhSorNPR1 shared 64.96% identity and 73.09% similarity to GhNPR1, a NPR1 ortholog originating from Gladiolus hybridus. A phylogenetic investigation of 32 NPR1s showed that LhSorNPR1 was phylogenetically closely related GdNPR1, a NPR1 orthologs originating from Musa spp. ABB (Fig. 4). Analysis of the intron/exon organization showed that LhSorNPR1 consisted of four exons and three introns. The intron/exon structure of NPR1 was conserved in Arabidopsis and rice as well as in other species, such as Theobroma cacao (Shi et al. 2010), Musa spp. ABB (Zhao et al. 2009), Malus hupehensis (Zhang et al. 2012), and soybean (Sandhu et al. 2009). Conservation of NPR1 in the intron/exon organization across different species (including lilies) indicated that the genomic structure of the NPR1 protein family was conserved. NPR1 orthologs share several domains, including the BTB/POZ domain, the ankyrin repeat, and the NLS, which were all found in LhSorNPR1. Disulfide bridges played a critical role in protein folding and structure stability maintenance (Patil et al. 2015). Previous studies demonstrated that five cysteine residues (cys82, cys150, cys155, cys160, and cys216) were important for the oligomer-monomer exchange of AtNPR1 (Mou et al. 2003; Tada et al. 2008). Among these five cysteine residues, cys82 and cys216 were found to be critical for the oligomer formation of NPR1 in Arabidopsis. Site-directed mutagenesis in either of these two cysteine residues caused monomerization and nuclear translocation of AtNPR1 to occur (Mou et al. 2003). In LhSorNPR1, four of these cys residues were conserved, cys216 being the exception. In point of fact, LhSorNPR1 was not the only NPR1 ortholog reported to have a nonconserved cys216. MhNPR1 (Zhang et al. 2012), GmNPR1-1 (Sandhu et al. 2009), GmNPR1-2, and MpNPR1 (Zhao et al. 2009) were all found to be inconsistent for cys216 residue after sequence alignment. Taking into consideration the importance of cys216, the inconsistence of this residue in different NPR1 orthologs indicated that additional cys residue(s) might be involved in facilitating oligomerization of NPR1 in cytoplasm. The SIM domain, which functioned in mediating conjugation of SUMO3 to the lysine residue(s) in AtNPR1 was conserved in LhSorNPR1 and OsNPR1. However, a IκB-like phosphodegron (S55/S59 in AtNPR1), whose phosphorylation is critical for the inhibition of AtNPR1 sumoylation was not conserved in LhSorNPR1. In light of the conservativeness of the SIM domain in NPR1 orthologues, it is likely that some other serine residues might play critical roles in regulation of sumoylation in LhSorNPR1.
The expression of a specific gene was mainly driven by the adjacent promoter. Analysis of the LhSorNPR1 promoter sequence uncovered CREs contributing to LhSorNPR1 expression regulation. The presence of the TCA element in the LhSorNPR1 promoter indicated that the LhSorNPR1 expression was responsive to SA, which was confirmed by qPCR analysis. At the same time, the LhSorNPR1 promoter also contains potential elements responsible for ethylene and abscisic acid. Although we did not find the motif for JA response, the LhSorNPR1 expression was responsive to the exogenous MeJA treatment (Fig. 6b). The presence of multiple phytohormone responsive elements in conjunction with the inducibility of LhSorNPR1 by MeJA, SA, and ETH revealed that the LhSorNPR1 expression was regulated by multiple hormones. Moreover, transcription factors, such as WRKY family proteins (Hwang and Hwang 2010), RAV1 proteins (Zhong et al. 2015), and ASF-1 proteins, have been reported to play important roles in NPR1 induction in different species. W boxes, especially the WBOXATNPR1 motif (Yu et al. 2001), within the LhSorNPR1 promoter, indicated that similar regulation mechanisms could exist for WRKY-involved expression regulation in Arabidopsis and lilies.
As a key node of SA-mediated signal transduction, NPR1 is a suitable candidate for genetic engineering in enhancing disease resistance in plants. Previous studies of carrots (Wally et al. 2009), citrus (Dutt et al. 2015), rice (Bai et al. 2011), and apples (Malnoy et al. 2007) have shown the feasibility of enhanced disease resistance by overexpressing NPR1. The NPR1 conferred resistance to Pseudomonas syringae and Peronospora parasitica was reported to exhibit a dosage-dependent manner (Cao et al. 1998) in transgenic Arabidopsis. Hence, it is possible that the overexpression of LhSorNPR1 could be used as a strategy to enhance disease resistance in lilies. Recently, the pivotal roles NPR1 played in salt and oxidative stress tolerance were also uncovered (Jayakannan et al. 2015). The findings of this study provide evidence of the importance of NPR1 in SA-mediated defense signaling. Moreover, our research and data on LhSorNPR1 could facilitate the genetic engineering of lilies in the future.
Acknowledgements
This research was supported by the Ningxia Agricultural Comprehensive Development Office (NTKJ2015-05-01), the National Natural Science Foundation of China (Grant No. 31201651), and the Hundred Talents Programs of Chinese Academy of Sciences (Nos. Y629721002 and 27Y127L41002).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Le Wang and Zhihong Guo have contributed equally to this work.
References
- Albagli O, Dhordain P, Deweindt C, Lecocq G, Leprince D. The BTB/POZ domain-a new protein-protein interaction motif common to DNA-binding and actin-binding proteins. Cell Growth Differ. 1995;6:1193–1198. [PubMed] [Google Scholar]
- Aravind L, Koonin EV. Fold prediction and evolutionary analysis of the POZ domain: structural and evolutionary relationship with the potassium channel tetramerization domain. J Mol Biol. 1999;285:1353–1361. doi: 10.1006/jmbi.1998.2394. [DOI] [PubMed] [Google Scholar]
- Bai W, Chern M, Ruan DL, Canlas PE, Sze-To WH, Ronald PC. Enhanced disease resistance and hypersensitivity to BTH by introduction of an NH1/OsNPR1 paralog. Plant Biotechnol J. 2011;9:205–215. doi: 10.1111/j.1467-7652.2010.00544.x. [DOI] [PubMed] [Google Scholar]
- Cao H, Bowling SA, Gordon AS, Dong XN. Characterization of an Arabidopsis mutant that Is nonresponsive to inducers of systemic acquired resistance. Plant Cell. 1994;6:1583–1592. doi: 10.1105/tpc.6.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong XN. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell. 1997;88:57–63. doi: 10.1016/S0092-8674(00)81858-9. [DOI] [PubMed] [Google Scholar]
- Cao H, Li X, Dong XN. Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. PNAS. 1998;95:6531–6536. doi: 10.1073/pnas.95.11.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke JD. Cetyltrimethyl ammonium bromide (CTAB) DNA miniprep for plant DNA isolation. Cold Spring Harb Protoc. 2009 doi: 10.1101/pdb.prot5177. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Delaney TP, Friedrich L, Ryals JA. Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. PNAS. 1995;92:6602–6606. doi: 10.1073/pnas.92.14.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despres C, DeLong C, Glaze S, Liu E, Fobert PR. The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell. 2000;12:279–290. doi: 10.1105/tpc.12.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutt M, Barthe G, Irey M, Grosser J. Transgenic citrus expressing an Arabidopsis NPR1 gene exhibit enhanced resistance against huanglongbing (HLB; Citrus Greening) PLoS ONE. 2015;10(9):e0137134. doi: 10.1371/journal.pone.0137134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu ZQ, Dong XN. Systemic acquired resistance: turning local infection into global defense. Annu Rev Plant Biol. 2013;64:839–863. doi: 10.1146/annurev-arplant-042811-105606. [DOI] [PubMed] [Google Scholar]
- Gaffney T, et al. Requirement of salicylic acid for the induction of systemic acquired resistance. Science. 1993;261:754–756. doi: 10.1126/science.261.5122.754. [DOI] [PubMed] [Google Scholar]
- Huang CJ, Liu YH, Yang KH, Chen CY. Physiological response of Bacillus cereus C1L-induced systemic resistance in lily against Botrytis leaf blight. Eur J Plant Pathol. 2012;134:1–12. doi: 10.1007/s10658-012-0013-6. [DOI] [Google Scholar]
- Hwang SH, Hwang DJ. Isolation and characterization of the rice NPR1 promoter. Plant Biotechnol Rep. 2010;4:29–35. doi: 10.1007/s11816-009-0116-5. [DOI] [Google Scholar]
- Jayakannan M, Bose J, Babourina O, Shabala S, Massart A, Poschenrieder C, Rengel Z. The NPR1-dependent salicylic acid signalling pathway is pivotal for enhanced salt and oxidative stress tolerance in Arabidopsis. J Exp Bot. 2015;66:1865–1875. doi: 10.1093/jxb/eru528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jue DW, Yang L, Shi C, Chen M, Yang Q. Cloning and characterization of a Solanum torvum NPR1 gene involved in regulating plant resistance to Verticillium dahliae. Acta Physiol Plant. 2014;36:2999–3011. doi: 10.1007/s11738-014-1671-0. [DOI] [Google Scholar]
- Katagiri F, Thilmony R, He SY. The Arabidopsis thaliana-pseudomonas syringae interaction. Arabidopsis Book. 2002;1:e0039. doi: 10.1199/tab.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesarwani M, Yoo JM, Dong XN. Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiol. 2007;144:336–346. doi: 10.1104/pp.106.095299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WK, Park MS, Hahm SS, Yu SH. Two new records of penicillium associated with blue moldy bulbs of lily in Korea. Mycobiology. 2006;34:176–179. doi: 10.4489/MYCO.2006.34.4.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Henanff G, et al. Vitis vinifera VvNPR1.1 is the functional ortholog of AtNPR1 and its overexpression in grapevine triggers constitutive activation of PR genes and enhanced resistance to powdery mildew. Planta. 2011;234:405–417. doi: 10.1007/s00425-011-1412-1. [DOI] [PubMed] [Google Scholar]
- Lecomte C, Alabouvette C, Edel-Hermann V, Robert F, Steinberg C. Biological control of ornamental plant diseases caused by Fusarium oxysporum: a review. Biol Control. 2016;101:17–30. doi: 10.1016/j.biocontrol.2016.06.004. [DOI] [Google Scholar]
- Liu YG, Chen Y. High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques. 2007;43:649. doi: 10.2144/000112601. [DOI] [PubMed] [Google Scholar]
- Malnoy M, Jin Q, Borejsza-Wysocka EE, He SY, Aldwinckle HS. Overexpression of the apple MpNPR1 gene confers increased disease resistance in malus x domestica. Mol Plant Microbe Interact. 2007;20:1568–1580. doi: 10.1094/MPMI-20-12-1568. [DOI] [PubMed] [Google Scholar]
- Metraux JP, et al. Increase in salicylic acid at the onset of systemic acquired resistance in cucumber. Science. 1990;250:1004–1006. doi: 10.1126/science.250.4983.1004. [DOI] [PubMed] [Google Scholar]
- Mou Z, Fan WH, Dong XN. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell. 2003;113:935–944. doi: 10.1016/S0092-8674(03)00429-X. [DOI] [PubMed] [Google Scholar]
- Patil NA, Tailhades J, Hughes RA, Separovic F, Wade JD, Hossain MA. Cellular disulfide bond formation in bioactive peptides and proteins. Int J Mol Sci. 2015;16:1791–1805. doi: 10.3390/ijms16011791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CM, Van Loon L. NPR1: the spider in the web of induced resistance signaling pathways. Curr Opin Plant Biol. 2004;7:456–464. doi: 10.1016/j.pbi.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Rasmussen JB, Hammerschmidt R, Zook MN. Systemic induction of salicylic acid accumulation in cucumber after inoculation with Pseudomonas syringae pv syringae. Plant Physiol. 1991;97:1342–1347. doi: 10.1104/pp.97.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochon A, Boyle P, Wignes T, Fobert PR, Despres C. The coactivator function of Arabidopsis NPR1 requires the core of its BTB/POZ domain and the oxidation of C-terminal cysteines. Plant Cell. 2006;18:3670–3685. doi: 10.1105/tpc.106.046953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals J, et al. The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor I kappa B. Plant Cell. 1997;9:425–439. doi: 10.1105/tpc.9.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A, et al. Transcriptional regulator NPR1 enable dynamic but tight control of plant immune responses. Cell Host Microbe. 2015;18:169–182. doi: 10.1016/j.chom.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu D, Tasma IM, Frasch R, Bhattacharyya MK. Systemic acquired resistance in soybean is regulated by two proteins, Orthologous to Arabidopsis NPR1. BMC Plant Biol. 2009;9:105. doi: 10.1186/1471-2229-9-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Maximova SN, Liu Y, Verica J, Guiltinan MJ. Functional analysis of the Theobroma cacao NPR1 gene in Arabidopsis. BMC Plant Biol. 2010;10:248. doi: 10.1186/1471-2229-10-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn KH, Lee SC, Jung HW, Hong JK, Hwang BK. Expression and functional roles of the pepper pathogen-induced transcription factor RAV1 in bacterial disease resistance, and drought and salt stress tolerance. Plant Mol Biol. 2006;61:897–915. doi: 10.1007/s11103-006-0057-0. [DOI] [PubMed] [Google Scholar]
- Spoel SH, Mou Z, Tada Y, Spivey NW, Genschik P, Dong XN. Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell. 2009;137:860–872. doi: 10.1016/j.cell.2009.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan T, Kumar KRR, Meur G, Kirti PB. Heterologous expression of Arabidopsis NPR1 (AtNPR1) enhances oxidative stress tolerance in transgenic tobacco plants. Biotechnol Lett. 2009;31:1343–1351. doi: 10.1007/s10529-009-0022-5. [DOI] [PubMed] [Google Scholar]
- Tada Y, et al. Plant immunity requires conformational changes of NPR1 via S-nitrosylation and thioredoxins. Science. 2008;321:952–956. doi: 10.1126/science.1156970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Vernooij B, et al. Salicylic acid Is not the translocated signal responsible for inducing systemic acquired resistance but is required in signal transduction. Plant Cell. 1994;6:959–965. doi: 10.1105/tpc.6.7.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wally O, Jayaraj J, Punja ZK. Broad-spectrum disease resistance to necrotrophic and biotrophic pathogens in transgenic carrots (Daucus carota L.) expressing an Arabidopsis NPR1 gene. Planta. 2009;231:131–141. doi: 10.1007/s00425-009-1031-2. [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature. 2001;414:562–565. doi: 10.1038/35107108. [DOI] [PubMed] [Google Scholar]
- Yu DQ, Chen CH, Chen ZX. Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell. 2001;13:1527–1539. doi: 10.1105/tpc.13.7.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JY, Qiao YS, Lv D, Gao ZH, Qu SC, Zhang Z. Malus hupehensis NPR1 induces pathogenesis-related protein gene expression in transgenic tobacco. Plant Biology. 2012;14:46–56. doi: 10.1111/j.1438-8677.2011.00483.x. [DOI] [PubMed] [Google Scholar]
- Zhang JY, Qu SC, Qiao YS, Zhang Z, Guo ZR. Overexpression of the Malus hupehensis MhNPR1 gene increased tolerance to salt and osmotic stress in transgenic tobacco. Mol Biol Rep. 2014;41:1553–1561. doi: 10.1007/s11033-013-3001-9. [DOI] [PubMed] [Google Scholar]
- Zhang YB, Wang YJ, Meng J, Xie ZK, Wang RY, Kutcher HR, Guo ZH. Development of an immunochromatographic strip test for rapid detection of lily symptomless virus. J Virol Methods. 2015;220:13–17. doi: 10.1016/j.jviromet.2015.03.021. [DOI] [PubMed] [Google Scholar]
- Zhang YB, Wang YJ, Yang WR, Xie ZK, Wang RY, Kutcher HR, Guo ZH. A rapid immunochromatographic test to detect the lily mottle virus. J Virol Methods. 2015;220:43–48. doi: 10.1016/j.jviromet.2015.04.010. [DOI] [PubMed] [Google Scholar]
- Zhao JT, Huang X, Chen YP, Chen YF, Huang XL. Molecular cloning and characterization of an ortholog of NPR1 gene from Dongguan Dajiao (Musa spp. ABB) Plant Mol Biol Rep. 2009;27:243–249. doi: 10.1007/s11105-008-0074-z. [DOI] [Google Scholar]
- Zhong X, et al. The NPR1 homolog GhNPR1 plays an important role in the defense response of Gladiolus hybridus. Plant Cell Rep. 2015;34:1063–1074. doi: 10.1007/s00299-015-1765-1. [DOI] [PubMed] [Google Scholar]