Figure 2.
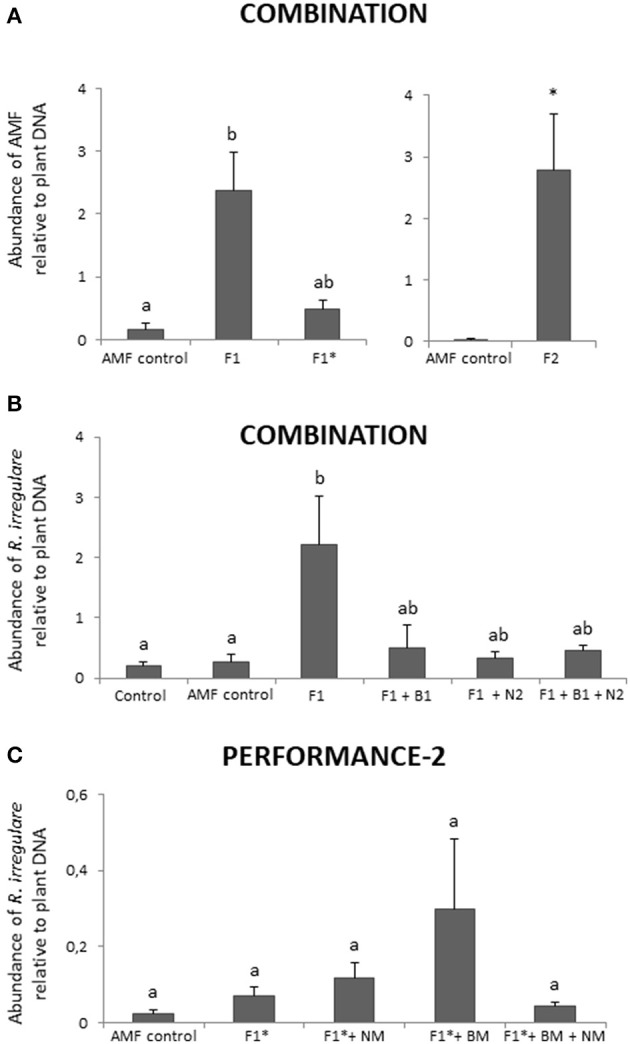
Abundance of Rhizoglomus irregulare in wheat roots in the COMBINATION (A,B) and PERFORMANCE-2 (C) field trials. (A) In the COMBINATION experiment, R. irregulare strain INOQ TOP was inoculated comparing high (F1) vs. low (F1*) dosages, with one of the treatments including the AMF strain SAF22 (F2). (B) In the same experiment, R. irregulare INOQ TOP (F1) was quantified in combination with bacteria, i.e., Pseudomonas protegens CHA0-Rif (B1), and nematodes, i.e., Heterorhabditis bacteriophora Andermatt (N2). (C) In the PERFORMANCE-2 experiment, R. irregulare INOQ TOP at the lower dosage (F1*) was used for the combination treatments with bacterial mixture (BM; i.e., P. protegens + Pseudomonas chlororaphis) and nematode mixture (NM; i.e., Heterorhabditis megidis + H. bacteriophora + Steinernema feltiae; for details see Figure S1). Control, non-inoculated control; AMF control, substrate control for AMF inoculation. R. irregulare was measured with quantitative PCR employing species-specific primers developed by Alkan et al. (2006) for INOQ TOP or their modified variants with enhanced specificity for SAF22 (Bender et al., unpublished). Bar graphs report mean normalized (R. irregulare relative to plant DNA) abundance (± SEM; COMBINATION, n = 4; PERFORMANCE-2, n = 7–9). Statistical analyses were performed on log-transformed data; asterisks and different letters indicate statistical significance at P < 0.05 for t-test and one-way ANOVA followed by the Tukey post-hoc test, respectively.
