Figure 1.
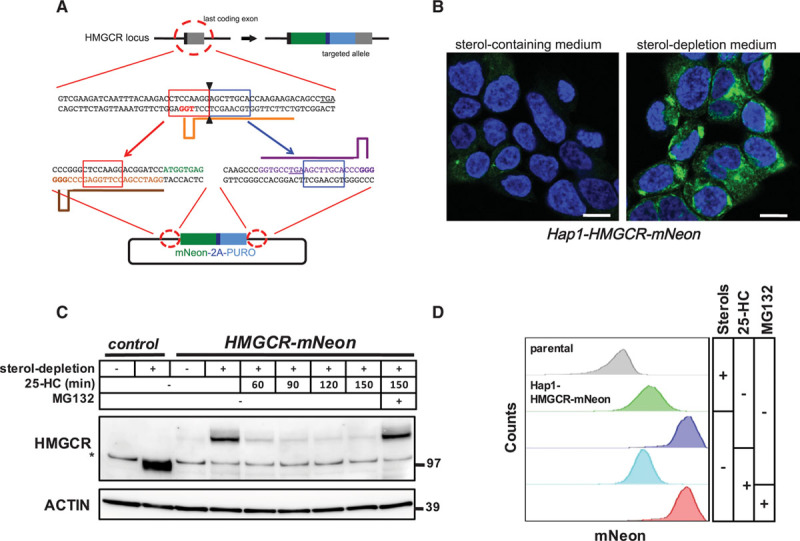
CRISPR/Cas9-mediated targeting of the endogenous HMGCR (3-hydroxy-3-methylglutaryl coenzyme A reductase) locus. A, Schematic illustration of CRISPR/Cas9-mediated targeting of the endogenous HMGCR locus for in-frame integration of mNeon-2A-PURO. Orange, brown, and purple bars correspond to the gRNA target sites. Red- and blue-boxed sequences indicate the microhomology sequences. The PAM sites are highlighted in bold, and the stop codons are underlined. B, Hap1-HMGCR-mNeon cells were grown on coverslips and cultured in sterol-containing or sterol-depletion medium for 24 h. Subsequently, cells were fixed, counterstained with 4′,6-diamidino-2-phenylindole, and imaged by confocal fluorescence microscopy. Representative images are shown (scale bar, 7.5 μm). C, Control Hap1 cells and Hap1-HMGCR-mNeon cells were grown in sterol-depletion or sterol-containing medium for 24 h, before treatment with 10 μmol/L 25-hydroxycholesterol (25-HC) and 25 μmol/L MG132 for the indicated time. Total cell lysates were immunoblotted as indicated. Immunoblot is representative of 3 independent experiments. The asterisk (*) indicates a nonspecific band. D, Hap1-HMGCR-mNeon cells were cultured as in (C) before addition of 10 μmol/L 25-HC and 25 μmol/L MG132 for 1 h after which the intensity of mNeon fluorescence was quantified by fluorescence-activated cell sorter analysis.
