Figure 2.
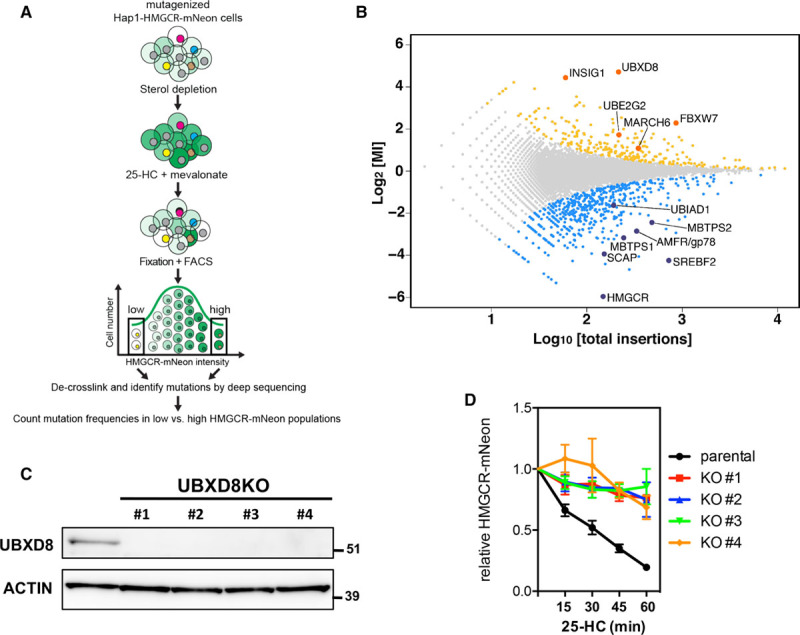
A mammalian haploid genetic screen identifies regulators of HMGCR (3-hydroxy-3-methylglutaryl coenzyme A reductase). A, Schematic illustration of the haploid genetic screen. A library of mutant Hap1-HMGCR-mNeon cells was generated by random, virus-mediated integration of a gene-trap cassette. To induce expression of HMGCR-mNeon, cells were cultured in sterol-depletion medium for 24 h. During the last 2 h, cells were treated with 10 μmol/L 25-hydroxycholesterol (25-HC) and 5 mmol/L mevalonate to initiate sterol-stimulated degradation of HMGCR. Genes that regulate HMGCR-mNeon abundance can be identified after cell sorting and recovery of the trapped loci. B, The log mutational index (MI) scores (see Materials and Methods in the online-only Data Supplement) were plotted against the number of trapped alleles per gene. Statistically significant hits (P<0.05) in the mNeonHIGH and mNeonLOW populations are indicated in yellow and blue, respectively (C). Total cell lysates of control and 4 independent UBXD8KO Hap1-HMGCR-mNeon cells were immunoblotted as indicated. Immunoblot is representative of 3 independent experiments. D, Control and UBXD8KO Hap1-HMGCR-mNeon cells were cultured in sterol-depletion medium for 24 h and subsequently treated with 10 μmol/L 25-HC for the indicated time. The intensity of HMGCR-mNeon was determined by fluorescence-activated cell sorter analysis, and the relative HMGCR-mNeon intensity is presented as mean±SD (n=3). KO indicates knockout; and UBXD8, ubiquitin regulatory X domain–containing protein 8.
