Abstract
Background:
We performed this meta-analysis to provide a comprehensive evaluation of the role of MicroRNA-210 (miR-210) expression on the overall survival (OS) rate of cancers.
Methods:
We searched for relevant available literatures on miR-210 and cancer until November 1st, 2016 on the databases PubMed, EMBASE, Cochrane Library, and Science Direct database. We calculated the pooled hazard ratio (HR) with 95% confidence intervals (CIs) for OS, which compared the high and low expression levels of miR-210 in patients of the available studies. Subgroup analysis was performed to evaluate the specific role of miR-210 in ethnicity and the type of cancers. Publication bias was evaluated using Begg funnel plots and Egger regression test.
Results:
Overall, 19 studies were involved in this meta-analysis. The result indicated that upregulated miR-210 might be associated with poor OS outcome in various carcinomas, with the pooled HR of 1.80 (95% CI: 1.29–2.51). When stratified by disease, significant results were detected in breast cancer (HR = 2.67, 95% CI: 1.24–5.76) and glioma (HR = 2.42, 95% CI: 1.32–4.43). Besides, in the subgroup analysis by ethnicity, significant results were detected only in Asian populations (HR = 2.14, 95% CI: 1.37–3.34).
Conclusion:
The present meta-analysis suggests that high expressed miR-210 is significantly associated with OS in cancer patients, which has the potential to be a prognostic marker in cancers.
Keywords: cancer, meta-analysis, MicroRNA-210, prognosis
1. Introduction
MicroRNAs (miRNA) are short sequences of nonprotein coding RNA, approximately 22 nucleotides in length. They could bind to specific regions of target mRNA transcripts, leading to mRNA degradation or translational repression.[1] In this way, miRNA have the potential to regulate various critical biological processes, such as cell cycle, apoptosis, and differentiation.[2] It is reported that miRNA dysregulation plays important roles in many diseases involving cancer.[3] Growing number of reports have shown that miRNAs are important regulators participating in tumorigenesis and tumor development.[4] Dysregulation of miRNAs can function as both tumor suppressors or oncogenes.[5,6] Recent studies have demonstrated that the expression of miRNA is found to correlate with the pathogenesis of many cancers, indicating that miRNAs may become valuable molecular markers for prognosis of tumors.[7]
MicroRNA-210 (miR-210) appears as an important regulator of cellular response to hypoxia, involving in cell cycle regulation, DNA damage repair, mitochondrial metabolism, tumor growth, apoptosis, and angiogenesis.[8] Its functional impact is reported both as an oncogene or a tumor suppressor, depending on tumor type. It has been proved that expression of miR-210 is associated with breast cancer (BC), renal cell carcinoma (RCC), nonsmall cell lung cancer (NSCLC), glioma, pediatric osteosarcoma, and so on.[9–13]
Recently, miR-210 was revealed to have prognostic value in some retrospective studies, which indicated that upregulation of miR-210 was correlated with poor prognosis in cancer patients. However, some other studies showed insignificant or opposite results.[22] In order to study the relationship between miR-210 and the survival of patients with cancers, we performed this meta-analysis. We discussed the possibility of using miR-210 as a prognostic marker in various carcinomas.
2. Materials and methods
2.1. Search strategy
A comprehensive search was applied to the following electronic databases PubMed, EMBASE, Cochrane Library, and Science Direct database to identify relevant studies, with the last search update on November 1st, 2016. The search strategy included the following keywords combined by “microRNA-210,” “miRNA-210,” “miR-210,” “cancer,” and “prognosis.” Furthermore, additional eligible studies were collected by a manual search for relevant researches from reference list.
2.2. Inclusion and exclusion criteria
Studies met the inclusion criteria: the patients in the eligible studies with any type of carcinoma; the expression of miR-210 in cancer tissues was measured; and the association between miR-210 expression levels and survival outcome was investigated. In addition, the following criteria were used to exclude published studies: case reports, letters, and reviews without original data; non-English papers; animal or laboratory studies; and lack of key information in the pooled hazard ratio (HR) and 95% confidence intervals (CIs). When overlapping data of the same patient population were included in more than 1 publication, only the most recent or complete study was used in this meta-analysis.
2.3. Data extraction
Two investigators (YL and YW) independently extracted the data with a standard protocol, and the result was reviewed by a 3rd investigator (XL). We extracted the following information: first author's surname, year of publication, type of disease, ethnicity, number of patients, miR-210 assay, survival analysis, follow-up time, and HR of miR-210 expression for overall survival (OS) with corresponding 95% CIs. Disagreements were discussed by other authors and resolved by consensus.
2.4. Statistical analysis
The statistical analyses were conducted using STATA software (version 12.0; StataCorp LP, College Station, TX). The pooled HR was calculated using a fixed-effects model or random-effects model to evaluate the relationship between miR-210 expression and OS. We evaluated the study heterogeneity analysis by I2 statistics.[14] The random effects model was used when an obvious heterogeneity was observed among the included studies (I2 > 50), otherwise the fixed effects model was selected to perform the meta-analysis. According to different diseases and ethnicity, subgroup analysis was further performed to minimize the influence of heterogeneity based on similar characteristics. Sensitivity analysis was carried out by excluding one single study one by one to check the stability and reliability on the recalculated HRs by repeating the meta-analysis. In addition, publication bias was estimated using Begg funnel plots and Eegg linear regression test, and P values were deemed as a significantly selective bias when less than 0.05.
3. Results
3.1. Studies characteristics
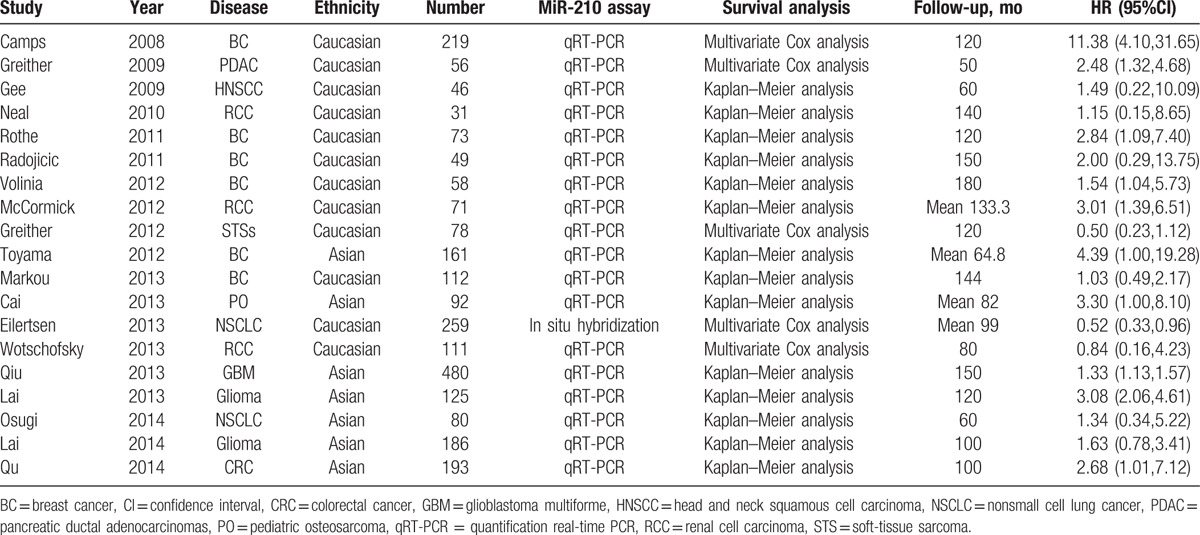
As is shown in Fig. 1, 339 records for miR-210 and cancer were retrieved. After screening titles and abstracts of identified articles, 275 were excluded because they were not related to the current study. Furthermore, 7 candidate studies were excluded because they were not giving available data. Finally, 19 studies were included in this meta-analysis, which were published between March 2008 and March 2015.[9–13,15–28] Among these previous studies, the types of cancers in these studies included BC, RCC, NSCLC, glioma, and so on. Besides, 18 studies used quantification real-time PCR to measure the expression level of miR-210, and only one used the method of in situ hybridization. In addition, 12 studies were conducted on Caucasian population, and the remaining 7 studies were on Asian population. In each study, the cut-off values of miR-210 appeared to be different. The main characteristics of eligible studies were summarized in Table 1.
Figure 1.
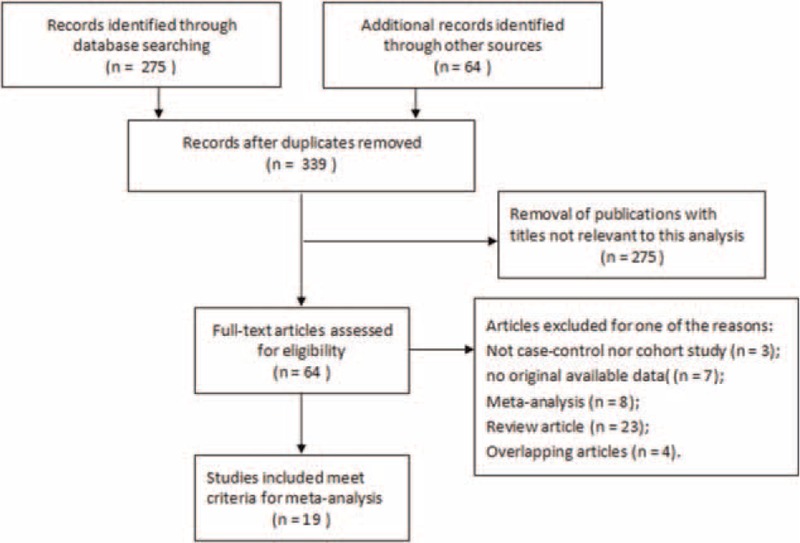
Flow diagram of the study selection process.
Table 1.
Characteristics of studies included in the meta-analysis.
3.2. Quantitative synthesis results
In this meta-analysis, we included 19 studies, a total of 2480 cases of patients with various carcinomas. The random-effect model was used to calculate the pooled HR with corresponding 95% CIs, because of the obvious between-study heterogeneity among these studies (I2 = 72.9%). The result indicated that upregulated miR-210 might be associated with poor OS outcome in various carcinomas, with the pooled HR of 1.80 (95% CI: 1.29–2.51) (Fig. 2). In the stratified analysis by disease, significant results were detected in BC (HR = 2.67, 95% CI: 1.24–5.76) and glioma (HR = 2.42, 95% CI: 1.32–4.43). But, in the stratified analysis by disease restricted to RCC and NSCLC, the results were not significantly different (Fig. 3A). Moreover, when stratified by ethnicity, significant results were detected only in Asian populations (HR = 2.14, 95% CI: 1.37–3.34) (Fig. 3B). Therefore, this meta-analysis indicated that high expression of miR-210 might predict poor survival in various carcinomas, such as BC and glioma.
Figure 2.
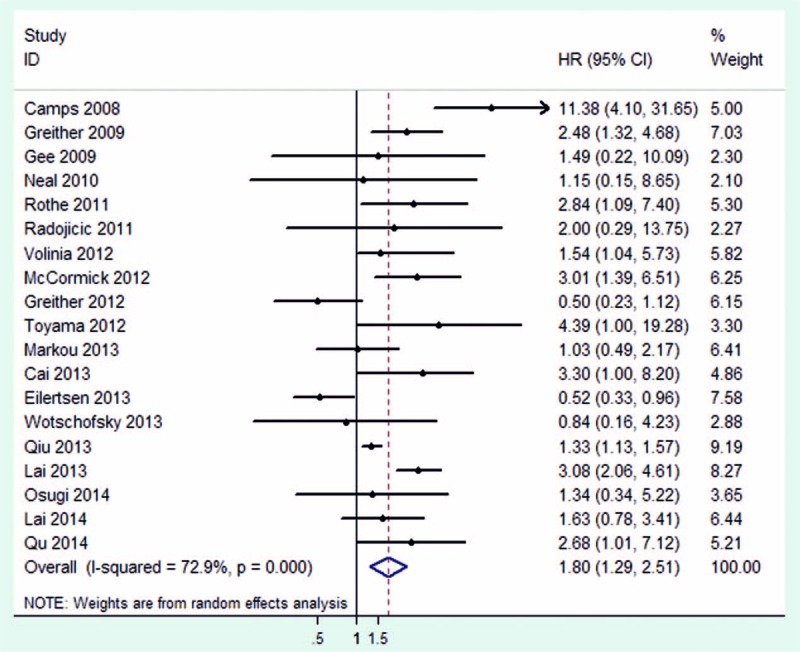
Pooled HR summary for OS with miR-210 expression in various carcinoma. HR = hazard ratio, miR-210 = MicroRNA-210, OS = overall survival.
Figure 3.
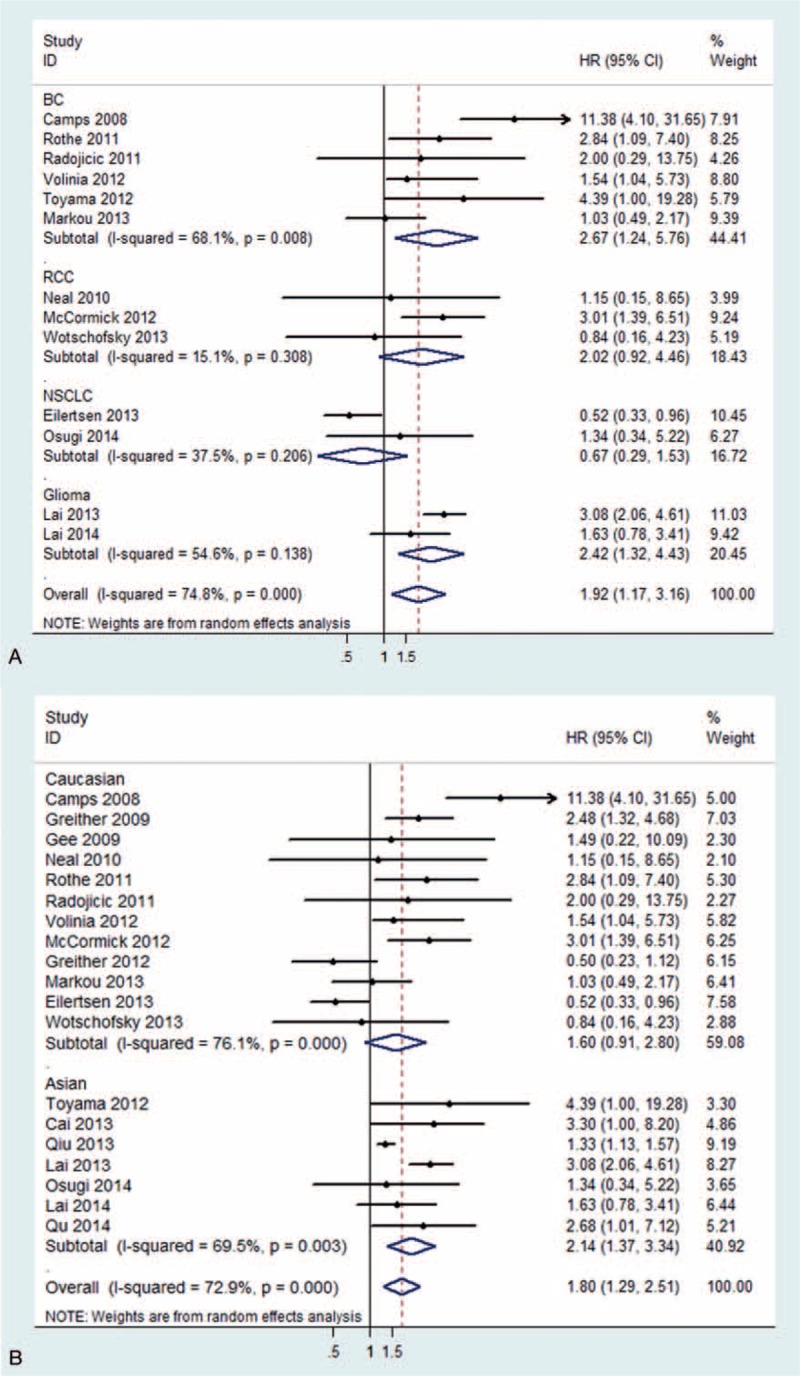
Pooled HR summary for OS with miR-210 expression in subgroup analysis. (A) Stratified by different diseases; (B) stratified by ethnicity. HR = hazard ratio, miR-210 = MicroRNA-210, OS = overall survival.
3.3. Sensitivity analysis
The sensitivity analysis on association between the expression levels of miR-210 and OS in cancer patients was listed in Fig. 4, suggesting that the pooled HR or the 95% CIs were not significantly influenced. Therefore, the sensitivity analysis revealed that our results were robust and reliable.
Figure 4.
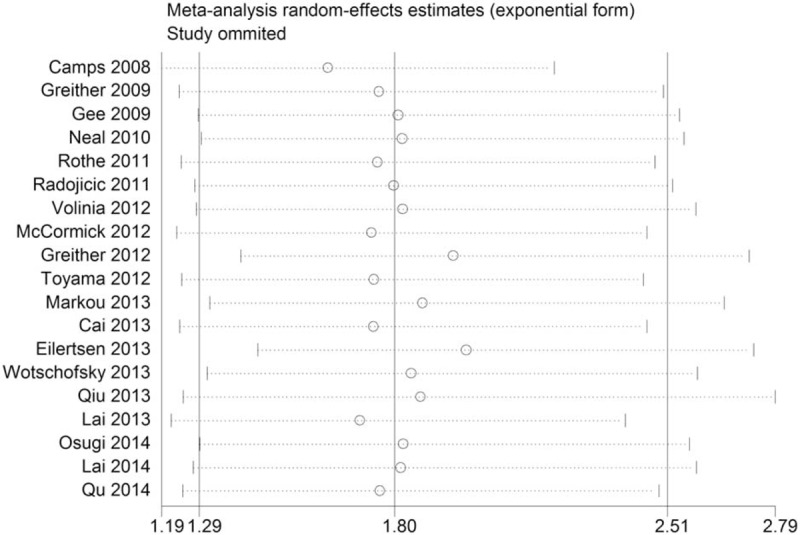
Sensitivity analysis of the association between high expression of miR-210 and the OS in various carcinomas. miR-210 = MicroRNA-210, OS = overall survival.
3.4. Publication bias
The Begg funnel plot and Eegg test was used to evaluate the publication bias of the literatures, which did not show any evidence of obvious asymmetry (P = .972) (Fig. 5).
Figure 5.
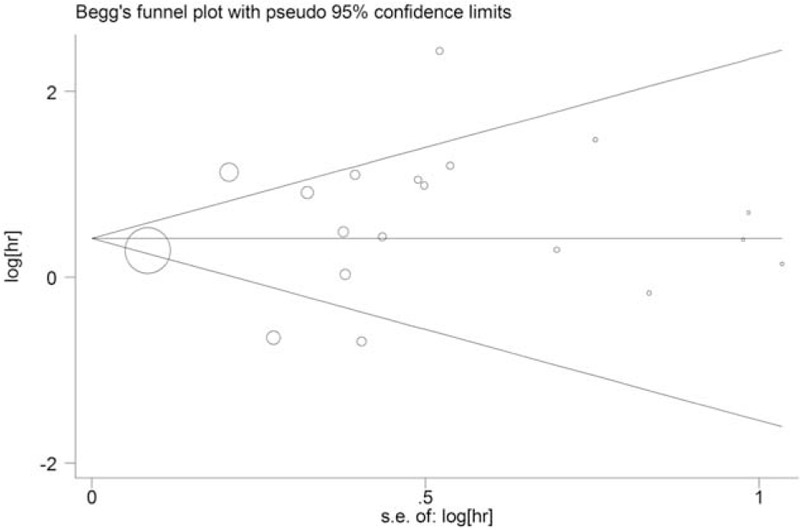
Begg funnel plot of publication bias test in various carcinomas.
4. Discussion
As one of small regulatory RNA molecules, MiRNA can modulate the expression of their target genes. In addition, miRNAs play vital roles in the physiological and pathological process, including cell proliferation, apoptosis, differentiation, and stress response.[29] Cancer is a multistep process in which normal cells undergo genetic damage along with hereditary susceptibility, which leads to the transformed phenotype characterized by deregulated gene expression and abnormal cell proliferation. During this transversion, there are many distinctive capacities such as self-sufficiency in growth, evasion of apoptosis, limitless replicative potential, growth inhibitory signals, sustained angiogenesis, and tissue invasion and metastasis in certain cancer cell.[30] Not surprizingly, a number of miRNAs have been implicated in cancer, in view of the obvious impact of miRNAs on gene expression. The importance of miRNAs in solid tumors and their potential utility as prognostic factors have become apparent.
MiR-210 is the miRNA most frequently associated with hypoxia which is a common feature of tumors.[31] Recent studies have demonstrated that expression of miR-210 was highly upregulated in hypoxic cells. It is located in an intron of a noncoding RNA, which plays important roles in cell survival and tumor initiation. It has been reported that its expression is elevated in various cancers involving BC, osteosarcoma, glioblastoma, head and neck cancer, NSCLC, RCC, and other solid tumors.[9–13] A growing number of studies found that miR-210 could function as a prognostic marker. Among them, Toyama et al[9] found low miR-210 expression experienced significantly better disease-free and OS than those with high miR-210 expression in BC, which could be an independent factor. Moreover, study from Cai et al[13] suggested that miR-210 upregulation showed a strong correlation with tumor aggressive progression of pediatric osteosaroma and could help prognostic screening of patients with this malignancy. However, some studies made the opposite results. Eilertsen et al[10] found that low expression of miR-210 was significantly and independently associated with a worse prognosis. Studies from Wotschofsky et al[21] identified that the expression of miR-210 could not be a predictive factor in RCC by the methods of multivariate Cox regression. Whether miR-210 acted as a tumor suppressor or an oncogene remained elusive. Thus, we conducted this meta-analysis to investigate the relationship between miR-210 expression and the survival in patients with cancer. Meanwhile, we also discussed the possibility of using miR-210 as a prognostic marker in various carcinomas.
In summary, the outcomes of previous studies depicting the association between high expression of miR-210 and the OS in various carcinomas remained inconclusive and controversial. The causes of these conflicts among them might be the various ethnicities, the relatively small sample size of each single study, the different kinds of diseases and the different methodologies in these published studies. However, since then, no meta-analysis explored such relationship. Recently, as the relevant studies published about association between expression levels of miR-210 and poor OS in various carcinomas has continued to increase, our meta-analysis was the first systematic evaluation to conduct a comprehensive evaluation of such relationship.
We performed this meta-analysis to illustrate controversial conclusions to make reliability of our meta-analysis and provide the most comprehensive understanding of such associations by different subgroup analysis. Accordingly, we took advantage of meta-analysis to explain this possible association. The elevated miR-210 expression level might be associated with poor OS outcome in various carcinomas. In addition, when stratified by different types of tumor, statistically significant increased the poor OS was shown in BC and glioma. Different kinds of tumor had their own various unique characteristics, which might lead to different statistical results. Besides, in the subgroup analysis by ethnicity, significant results were found only in Asian populations. Although the exact mechanism was unknown, it was likely that changes of different expression levels of miRNA in various tumors might have an important influence in different ethnic groups with various genetic backgrounds. In the current meta-analysis, our results revealed high expression of miR-210 might predict poor survival in various tumor, especially among BC and glioma.
The present meta-analysis found that elevated expression of miR-210 did indeed predict poor survival in patients with a variety of carcinoma. However, there are several limitations in our meta-analysis. First, though 19 studies were included in this meta-analysis, only a few articles were searched in a certain tumor, which might weaken the reliability of our results. Thus, more well-designed clinical studies with large cases of each specific cancer should be performed in the future to validate the relationship between miR-210 expression level and prognosis of cancerous patients. Second, the cut-off value of miR-210 should better be clearly defined. Thereby, a more precise analysis would have been performed if more clear and unified cut-off value was defined. What is more, the obvious heterogeneity existed in our meta-analysis and probably due to the difference in races, the tumor types, the disease stages, the cut-off value of miR-210 expression, the duration of follow-up, and so on. In addition, only studies with full text from English databases were selected, which might have led to publication. Furthermore, we did not have enough data in all studies published to adjust estimates by other influencing factors, including gender, age, lifestyle, and so on in the present meta-analysis. Nevertheless, only Caucasian and Asian populations were investigated in these majority studies, suggesting the results might exist some merits. As a consequence, more researches should pay close attention to the influence of various factors to guaranty these results more credible in the future.
5. Conclusion
In this meta-analysis, we drew the conclusion that high expressed miR-210 was significantly associated with poor survival in patients with various types of carcinoma, especially in BC and glioma. Besides, more studies should be carried out to confirm these findings in the future. MiR-210 might be the potential to be a prognostic factor in various carcinomas.
Acknowledgments
The authors thank Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), Program for Provincial Initiative Program for Excellency Disciplines of Jiangsu Province, National Natural Science Foundation of China (81672531, 81372757), and Six talent peak project in jiangsu province (WSN-011) for the support.
Footnotes
Abbreviations: BC = breast cancer, CI = confidence interval, HR = hazard ratio, miR-210 = MicroRNA-210, miRNA = MicroRNAs, NSCLC = nonsmall cell lung cancer, OS = overall survival, qRT-PCR = quantification real-time PCR, RCC = renal cell carcinoma.
YL, YW, and QX contributed equally to this work.
Funding/support: This study was supported by Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), Program for Provincial Initiative Program for Excellency Disciplines of Jiangsu Province, National Natural Science Foundation of China (81672531, 81372757), and Six talent peak project in jiangsu province (WSN-011).
The authors have no conflicts of interest to disclose.
References
- [1].Yang W, Lee DY, Ben-David Y. The roles of microRNAs in tumorigenesis and angiogenesis. Int J Physiol Pathophysiol Pharmacol 2011;3:140–55. Resource. [PMC free article] [PubMed] [Google Scholar]
- [2].Oliveto S, Mancino M, Manfrini N, et al. Role of microRNAs in translation regulation and cancer. World J Biol Chem 2017;8:45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kong YW, Ferland-McCollough D, Jackson TJ, et al. microRNAs in cancer management. Lancet Oncol 2012;13:e249–58. [DOI] [PubMed] [Google Scholar]
- [4].Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol 2012;6:590–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Liu J, Zheng M, Tang YL, et al. MicroRNAs, an active and versatile group in cancers. Int J Oral Sci 2011;3:165–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lovat F, Valeri N, Croce CM. MicroRNAs in the pathogenesis of cancer. Semin Oncol 2011;38:724–33. [DOI] [PubMed] [Google Scholar]
- [7].Fiorucci G, Chiantore MV, Mangino G, et al. Cancer regulator microRNA: potential relevance in diagnosis, prognosis and treatment of cancer. Curr Med Chem 2012;19:461–74. Resource. [DOI] [PubMed] [Google Scholar]
- [8].Fasanaro P, Greco S, Lorenzi M, et al. An integrated approach for experimental target identification of hypoxia-induced miR-210. J Biol Chem 2009;284:35134–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Toyama T, Kondo N, Endo Y, et al. High expression of microRNA-210 is an independent factor indicating a poor prognosis in Japanese triple-negative breast cancer patients. Jpn J Clin Oncol 2012;42:256–63. [DOI] [PubMed] [Google Scholar]
- [10].Eilertsen M, Andersen S, Al-Saad S, et al. Positive prognostic impact of miR-210 in non-small cell lung cancer. Lung Cancer 2014;83:272–8. [DOI] [PubMed] [Google Scholar]
- [11].Neal CS, Michael MZ, Rawlings LH, et al. The VHL-dependent regulation of microRNAs in renal cancer. BMC Med 2010;8:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lai NS, Dong QS, Ding H, et al. MicroRNA-210 overexpression predicts poorer prognosis in glioma patients. J Clin Neurosci 2014;21:755–60. [DOI] [PubMed] [Google Scholar]
- [13].Cai H, Lin L, Cai H, et al. Prognostic evaluation of microRNA-210 expression in pediatric osteosarcoma. Med Oncol 2013;30:499. [DOI] [PubMed] [Google Scholar]
- [14].Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ 2003;327:557–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].McCormick RI, Blick C, Ragoussis J, et al. miR-210 is a target of hypoxia-inducible factors 1 and 2 in renal cancer, regulates ISCU and correlates with good prognosis. Br J Cancer 2013;108:1133–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lai NS, Wu DG, Fang XG, et al. Serum microRNA-210 as a potential noninvasive biomarker for the diagnosis and prognosis of glioma. Br J Cancer 2015;112:1241–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gee HE, Camps C, Buffa FM, et al. hsa-mir-210 is a marker of tumor hypoxia and a prognostic factor in head and neck cancer. Cancer 2010;116:2148–58. [DOI] [PubMed] [Google Scholar]
- [18].Radojicic J, Zaravinos A, Vrekoussis T, et al. MicroRNA expression analysis in triple-negative (ER, PR and Her2/neu) breast cancer. Cell Cycle 2011;10:507–17. [DOI] [PubMed] [Google Scholar]
- [19].Camps C, Buffa FM, Colella S, et al. hsa-miR-210 Is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res 2008;14:1340–8. [DOI] [PubMed] [Google Scholar]
- [20].Markou A, Yousef GM, Stathopoulos E, et al. Prognostic significance of metastasis-related microRNAs in early breast cancer patients with a long follow-up. Clin Chem 2014;60:197–205. [DOI] [PubMed] [Google Scholar]
- [21].Wotschofsky Z, Busch J, Jung M, et al. Diagnostic and prognostic potential of differentially expressed miRNAs between metastatic and non-metastatic renal cell carcinoma at the time of nephrectomy. Clin Chim Acta 2013;416:5–10. [DOI] [PubMed] [Google Scholar]
- [22].Greither T, Wurl P, Grochola L, et al. Expression of microRNA 210 associates with poor survival and age of tumor onset of soft-tissue sarcoma patients. Int J Cancer 2012;130:1230–5. [DOI] [PubMed] [Google Scholar]
- [23].Greither T, Grochola LF, Udelnow A, et al. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int J Cancer 2010;126:73–80. [DOI] [PubMed] [Google Scholar]
- [24].Osugi J, Kimura Y, Owada Y., et al. Prognostic impact of hypoxia-inducible miRNA-210 in patients with lung adenocarcinoma. J Oncol 2015;2015: Article ID 316745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Qiu S, Lin S, Hu D, et al. Interactions of miR-323/miR-326/miR-329 and miR-130a/miR-155/miR-210 as prognostic indicators for clinical outcome of glioblastoma patients. J Transl Med 2013;11:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Rothe F, Ignatiadis M, Chaboteaux C, et al. Global microRNA expression profiling identifies MiR-210 associated with tumor proliferation, invasion and poor clinical outcome in breast cancer. PLoS One 2011;6:e20980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Qu A, Du L, Yang Y, et al. Hypoxia-inducible MiR-210 is an independent prognostic factor and contributes to metastasis in colorectal cancer. PLoS One 2014;9:e90952. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [28].Volinia S, Galasso M, Sana ME, et al. Breast cancer signatures for invasiveness and prognosis defined by deep sequencing of microRNA. Proc Natl Acad Sci U S A 2012;109:3024–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281–97. Resource. [DOI] [PubMed] [Google Scholar]
- [30].Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000;100:57–70. Resource. [DOI] [PubMed] [Google Scholar]
- [31].Hong L, Han Y, Zhang Y, et al. MicroRNA-21: a therapeutic target for reversing drug resistance in cancer. Expert Opin Ther Targets 2013;17:1073–80. [DOI] [PubMed] [Google Scholar]


