Fig. 1.
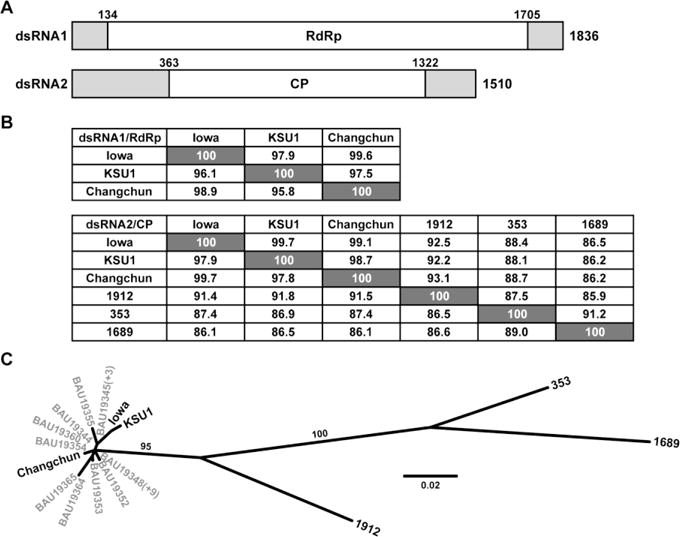
(A) Diagram of the genomic RNA plus strands of CSpV1-Iowa. The beginning and end of the long ORF encoding RdRp or CP are indicated by nt positions in each RNA. Terminal NTRs are shaded. (B) Pairwise alignment scores (%). Values at the lower left of each panel are for the protein-encoding RNA sequences of each genome segment; values at the upper right are for the deduced protein sequences. (C) Maximum-likelihood phylogenetic tree (substitution model, LG; gamma shape parameter, 0.751; proportion of invariant, 0.184) for the CP sequences of the indicated cryspovirus strains (see main text). Branch support values (%) are shown for only the two long branches. Some of the C. parvum-derived strains from Murakoshi et al. [18] share the same CP sequence, indicated by (+N) in the figure.
