Fig. 1.
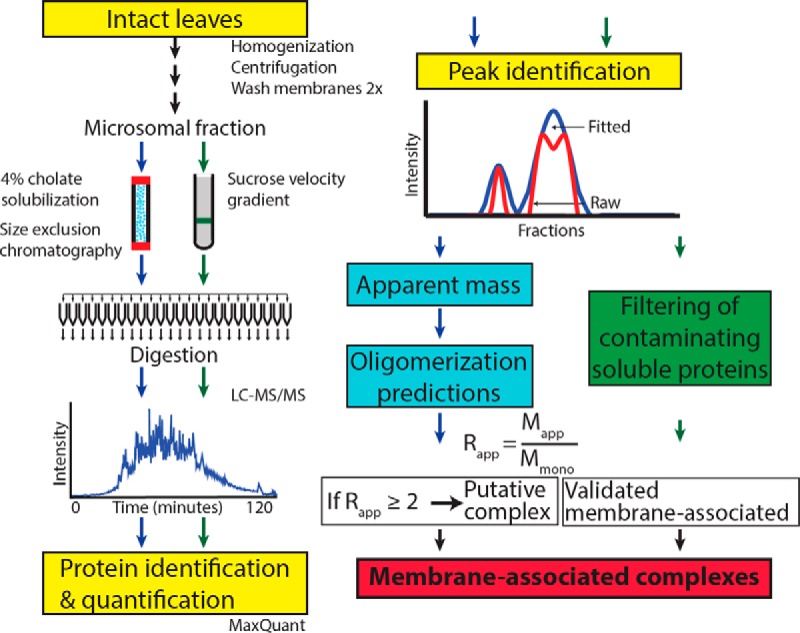
Workflow for a proteomic analysis of membrane-associated protein complexes. Arabidopsis leaves were homogenized and a crude microsomal fraction was isolated. Microsomal proteins were solubilized in cholate and resolved by size exclusion chromatography. Fractions were collected, digested, and analyzed by LC-MS/MS. MaxQuant was used to identify the peptides and generate XIC abundance profiles. Peaks in the elution profiles were identified using Gaussian fitting. From the SEC profiles protein oligomerization predictions were made by calculating the ratio (Rapp) of the apparent mass (Mapp) determined by SEC to the monomer mass (Mmono) of the protein. Proteins with a Rapp ≥2 were predicted to form a putative complex. Contaminating soluble proteins were removed from the data set based their inability to sufficiently penetrate a sucrose gradient following ultracentrifugation (green box).
