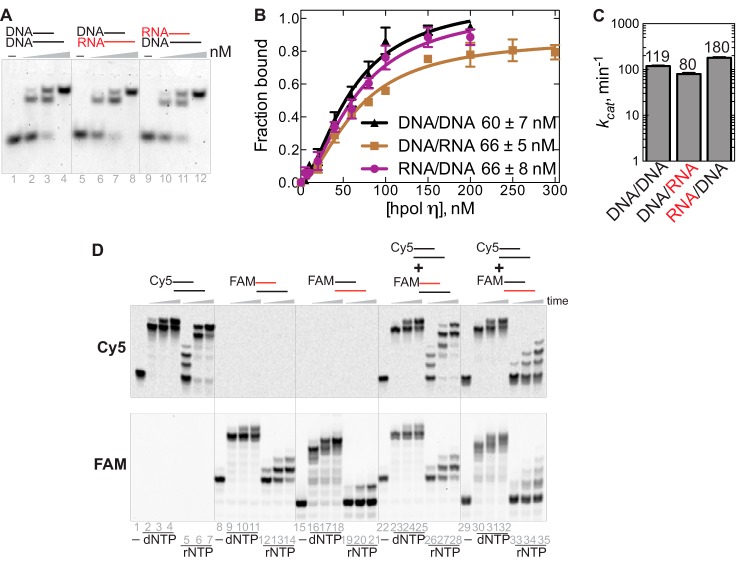
Figure 5.
Substrate binding and competition assay for hpol η. A, annealed substrates DNA/DNA, DNA/RNA, and RNA/DNA (20 nm) were incubated with 0, 50, 100, and 200 nm hpol η, respectively, at 30 °C for 30 min. B, titration of hpol η (0–300 nm) with the different substrates (20 nm). The quantified data were best fit with the equation, Y = Bmax × Xh/(Kdh + Xh) (in GraphPad Prism). The maximum fraction bound values, Bmax, were close to unity for all three substrates, and the Hill coefficients were 1.6 ± 0.2, 1.7 ± 0.2, and 1.7 ± 0.2 for DNA/DNA, DNA/RNA, and RNA/DNA complexes, respectively, indicative of multiple proteins binding with a molecule of DNA. The Kd value for each substrate is indicated in the figure. C, comparison of kcat values from steady-state kinetics for each substrate by hpol η. D, Cy5-DNA/DNA was mixed with FAM-RNA/DNA or FAM-DNA/RNA at a molar ratio of 1:1. hpol η (500 nm) and annealed substrates (5 μm) were reacted with the mixture of dNTPs or rNTPs (1 mm concentration of each nucleotide) for 5, 30, and 60 min. The top panel was scanned in the Cy5 channel, and the bottom panel was scanned in the FAM channel (GE Typhoon system).