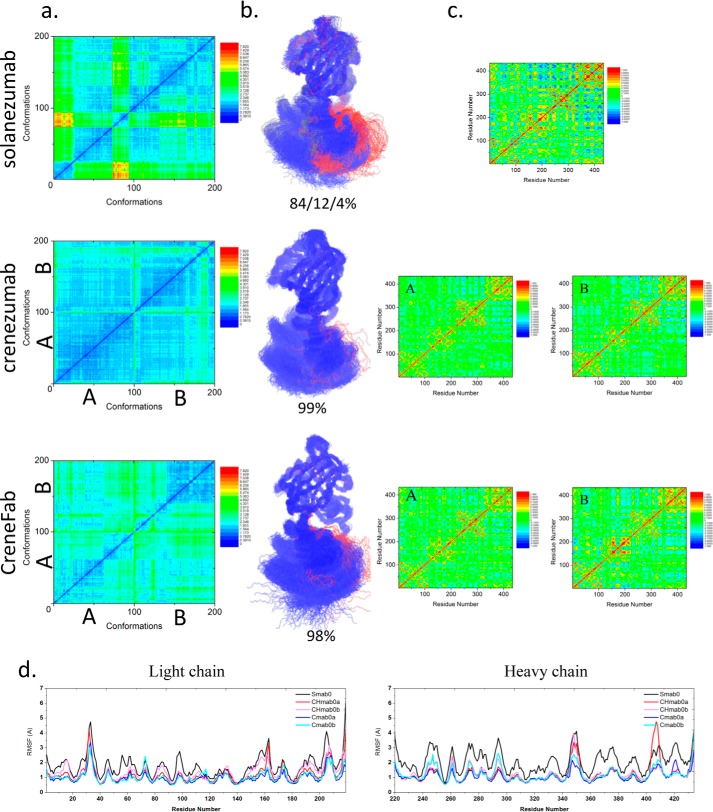
Figure 1.
The structure and dynamics of the three Fabs in their apo form suggest that when unbound to antigens, solanezumab is more conformationally diverse than the other two Fabs. a, 2D RMSDs of the structures sampled in the MD simulation. The structures of crenezumab and CreneFab are taken from PDB entries 4KMV (A) and 4KNA (B), respectively. b, cluster analysis, with backbone RMSD = 4 Å, used to define the cluster. Clusters are colored blue, red, and gray, respectively. c, motion correlation among the residues of the three Fabs. Residues with highly correlated and anti-correlated motion are red and blue, respectively. d, RMSFs of the three Fabs from five independent MD simulations. The RMSFs of Smab0, CHmab0a, CHmab0b, Cmab0a, and Cmab0b are colored black, red, pink, blue, and cyan, respectively.