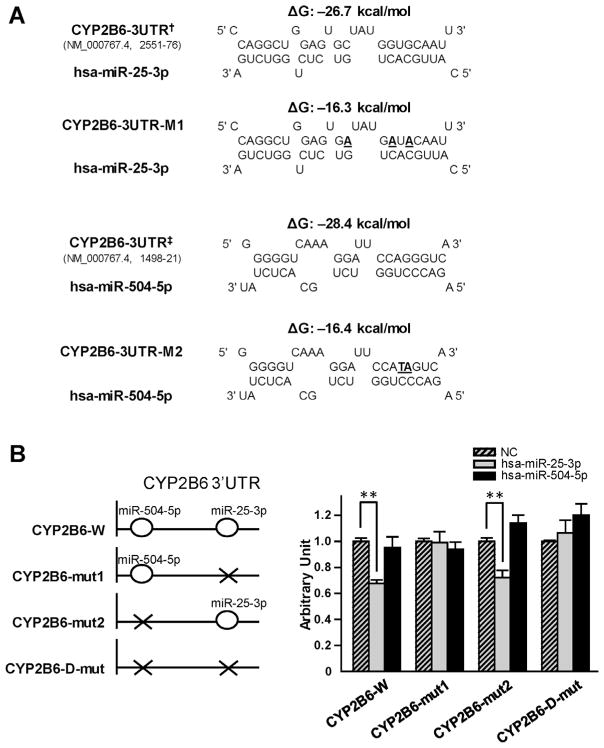
Fig. 2.
Hsa-miR-25-3p down-regulated luciferase reporter activities by targeting CYP2B6 3′-UTR. (A) Predicted hybrid complexes for hsa-miR-25-3p and hsa-miR-504-5p with sequences found in the wild-type CYP2B6 3′-UTR and with altered sequences (bold, underlined). † and ‡ indicated the nucleotide positions 2551–76 and 1498–21 at the CYP2B6 3′-UTR in NM_000767.4, counting from the transcription start site as nucleotide position 1. CYP2B6-3UTR-M1 and CYP2B6-3UTR-M2 indicated the mutated hsa-miR-25-3p and hsa-miR-504-5p targeting sequences based on the wild-type CYP2B6 3′-UTR, respectively. (B) The diagrams on the left depict the structures of the reporter gene constructs used in this study. CYP2B6-W construct contains the wild-type CYP2B6 3′-UTR. CYP2B6-mut1 and CYP2B6-mut2 constructs contain the 3′-UTR fragment with the CYP2B6-3UTR-M1 and CYP2B6-3UTR-M2 mutant sequences, respectively. The CYP2B6-D-mut contains the 3′-UTR with both the CYP2B6-3UTR-M1 and the CYP2B6-3UTR-M2 mutant sequences. The graph on the right shows that luciferase reporter gene activity in HEK 293T cells was suppressed by transfection with hsa-miR-25-3p mimics. The luciferase reporter plasmids (100 ng) were co-transfected into HEK 293T cells together with 50 nmol/L of miRNA negative controls (NC; striped bars), hsa-miR-25-3p (gray bars) or has-miR-504-5p mimics (black bars), respectively. Data are presented as mean ± S.D. (n = 9). **P < 0.001.