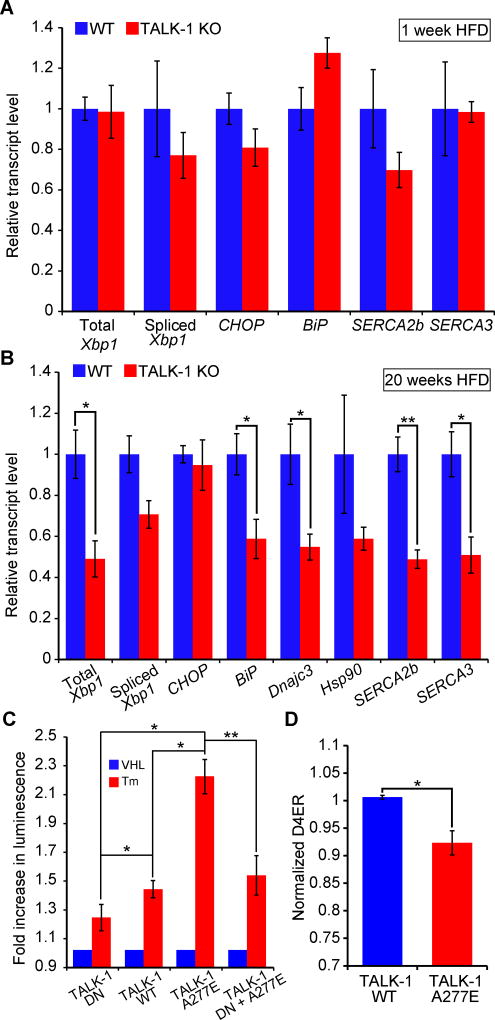
Figure 8. TALK-1 channel activity exacerbates ER stress.
(A) Reverse-transcribed RNA from islets isolated from wild-type (WT) and TALK-1 KO mice fed a HFD for 1 week was subjected to quantitative real-time PCR (qRT-PCR) to measure total Xbp1, spliced Xbp1, CHOP, BiP, Atp2a2 (SERCA2b), and Atp2a3 (SERCA3) expression (N = 4–5 mice per genotype). (B) Reverse-transcribed RNA from islets isolated from wild-type (WT) and TALK-1 KO mice fed a HFD for 20 weeks was subjected to qRT-PCR to measure total Xbp1, spliced Xbp1, CHOP, BiP, Dnajc3, Hsp90, Atp2a2 (SERCA2b), and Atp2a3 (SERCA3) expression (N = 3–4 mice per genotype). (C) INS-1 cells co-transfected with TALK-1 DN mutant, wild-type TALK-1 (WT), or TALK-1 A277E and an ATF6 promoter luciferase reporter (p5×ATF6-GL3) were treated with vehicle (VHL) (DMSO, 0.0125% v/v) or tunicamycin (Tm) (0.25 µg/mL) for 16–20 hours prior to cell lysis and luciferase assay (N = 4 independent experiments). (D) INS-1 cells were co-transfected with TALK-1 WT or TALK-1 A277E and pCMV-D4ER to measure basal Ca2+ER concentrations in 11 mM glucose (N = 4 independent experiments). Statistical significance was determined by Student’s t-test; *P<0.05, **P<0.01.