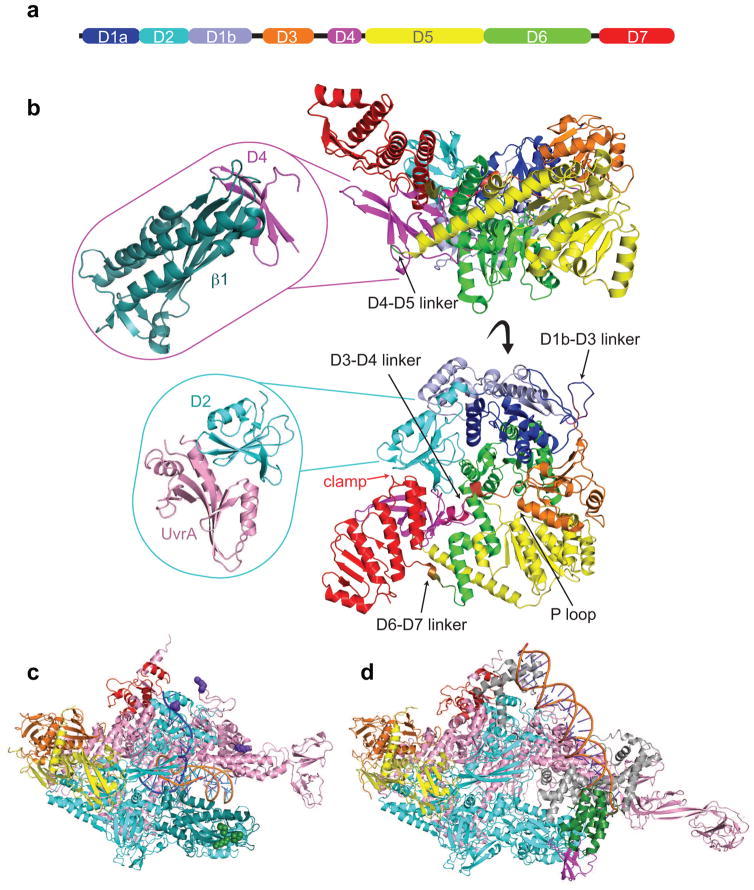
Figure 3. Structural features of key TCR players.
a) Schematic representation of Mfd domain architecture colored as follows: D1a in blue, D2 in cyan, D1b in slate, D3 in orange, RID in magenta, D5 in yellow, D6 in green, and D7 in red. The horizontal bar represents the 1148-residue primary sequence with colored blocks indicating domains and black lines highlighting flexible connecting linkers.
b) Top and side views showing an α carbon backbone ribbon representation of the E. coli apo Mfd structure (PDB ID 2EYQ) with domains colored as in (a). Top inset shows an α carbon ribbon representation of Thermus sp. core Mfd-RID/RNAP-β complex (PDB ID 3MLQ). Bottom inset shows an α carbon representation of E. coli core Mfd/UvrA complex (PDB ID 4DFC).
c) Overall architecture of Thermus thermophilus RNAP elongation complex (PDB ID 2O5I) with RNAP subunits colored as follows: α1 and α2 in orange and yellow, β in cyan, β′ in pink, ω in red. The N-terminal region of the β subunit interacting with Mfd-RID is colored in deep teal. Residues involved in direct contacts with Mfd-RID (the “IKE” motif) are shown as green spheres (β subunit I108, K109, and E110). Residues involved in direct contacts with UvrD are shown as purple spheres (β subunit K781, β′ subunit K28 and R67).
d) Overall architecture of Thermus aquaticus transcription initiation complex bound to CarD (PDB ID 4XLS). RNAP subunits are colored as in (c) and the σ factor in gray. The CarD-RID is magenta and CarD-CTD is green.