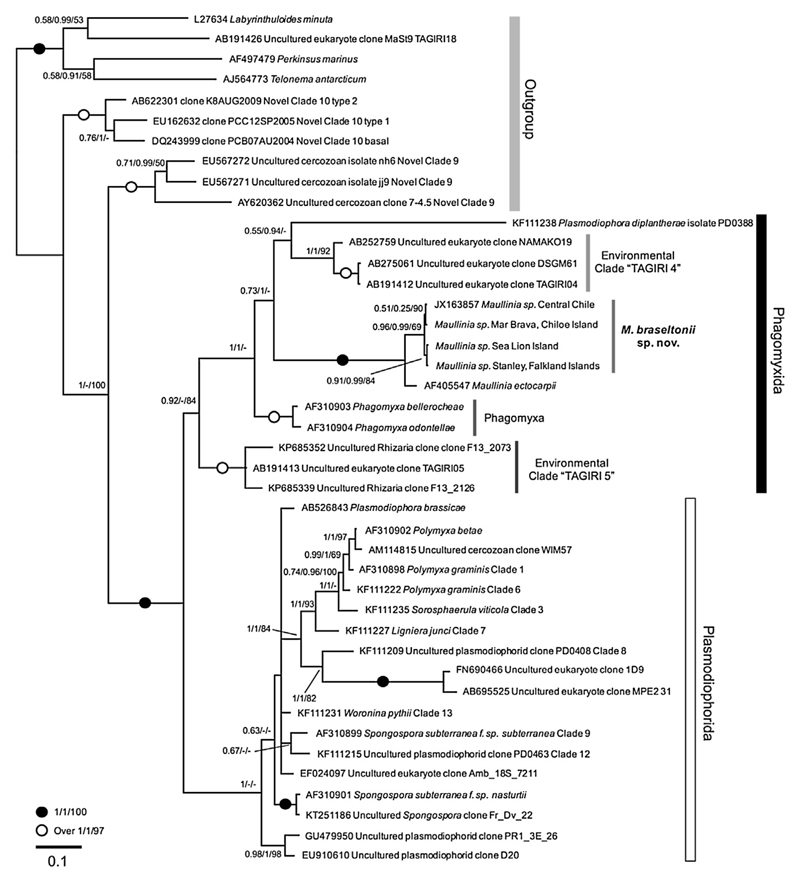
Figure 5.
Bayesian analyses of M. braseltonii and all available phytomyxid sequences (from known species and environmental 18S rDNA clades). The tree contains a total of 43 sequences and 1002 positions. Support values given are posterior probabilities (MrBayes)/χ2 support values (PhyML)/bootstrap support (RAxML). MrBayes settings: chain length 1.000.000, subsample frequency 1.000, burn in of 10%. The scale bar indicates the number of substitutions per site. The accompanying PhyML (Supplementary Fig. 2) and RAxML (Supplementary Fig. 3) trees are provided in the Supplement. The genus Maullinia is well supported and M. braseltonii and M. ectocarpii form distinct branches on the tree. Phytomyxea (Phagomyxida and Plasmodiophorida) form a well-supported, monophyletic clade.