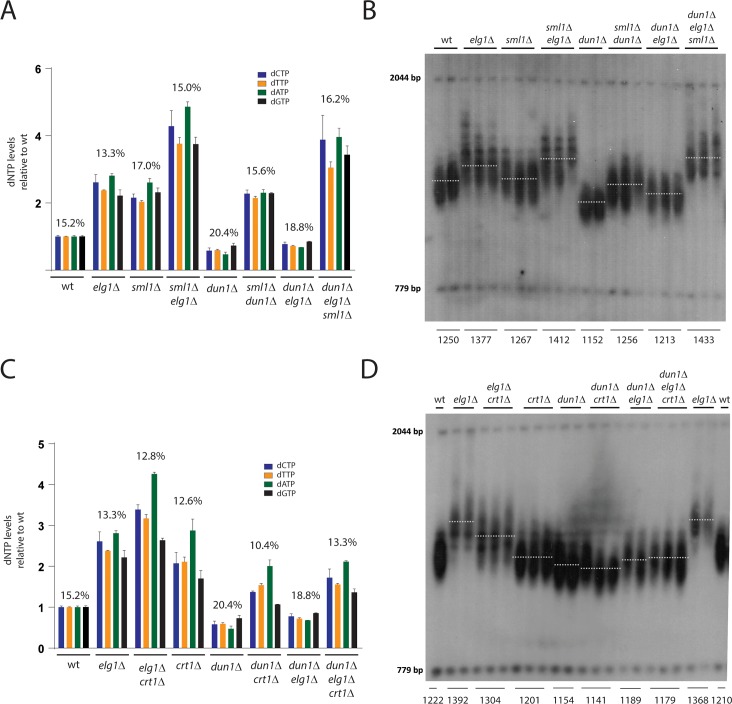
Fig 5. The Mec1ATR-Dun1 pathway promotes telomere over-elongation via Rnr1 activation through Sml1 degradation.
A) dNTP pools and dGTP ratios (in %) were measured in the indicated strains. dun1Δ mutants show low dNTP levels, which can be elevated above wild type levels by a co-deletion of SML1. In sml1Δ elg1Δ dun1Δ mutants dNTP levels are elevated approximately 4 fold relative to wild type. Mean values +/-SEM are indicated. All values are shown as fold change over the individual dNTP levels of the wild type which were taken from the experiment in Fig 1C (set to 1). n(wt) = 9; n(elg1Δ) = 2; n(sml1Δ) = 2; n(sml1Δ elg1Δ) = 2; n(dun1Δ) = 2; n(sml1Δ dun1Δ) = 2; n (dun1Δ elg1Δ) = 2; n(sml1Δ dun1Δ elg1Δ) = 2. B) Telomere length analysis by Southern blotting. A deletion of DUN1 results in short telomeres, which only slightly become elongated by a co-deletion of the long TLM gene ELG1. The simultaneous deletion of SML1 restores telomere length in dun1Δ mutants and rescues the over-elongation of telomeres in dun1Δ elg1Δ double mutants. Represented are biological replicates of the indicated strains after approximately 200 generations. C) dNTP pools and dGTP ratios (in %) were measured in the indicated strains. dun1Δ mutants show low dNTP levels, which can be elevated above wild type levels by a deletion of CRT1 or simultaneous deletions of CRT1 and ELG1. Mean values +/-SEM are indicated. All values are shown as fold change over the individual dNTP levels of the wild type which were taken from the experiment in Fig 1C (set to 1). The dNTP levels of elg1Δ and dun1Δ mutants were taken from the experiment in (A) and are shown as a comparison. n(wt) = 9; n(elg1Δ) = 2; n(elg1Δ crt1Δ) = 2; n(crt1Δ) = 2; n(dun1Δ) = 2; n(dun1Δ crt1Δ) = 2; n(dun1Δ elg1Δ) = 2; n(dun1Δ elg1Δ crt1Δ) = 2. D) Telomere length analysis by Southern blotting. A co-deletion of CRT1 does not restore telomere length in dun1Δ mutants nor does it rescue the over-elongation of telomeres in dun1Δ elg1Δ double mutants. Represented are biological replicates of the indicated strains after approximately 200 generations.