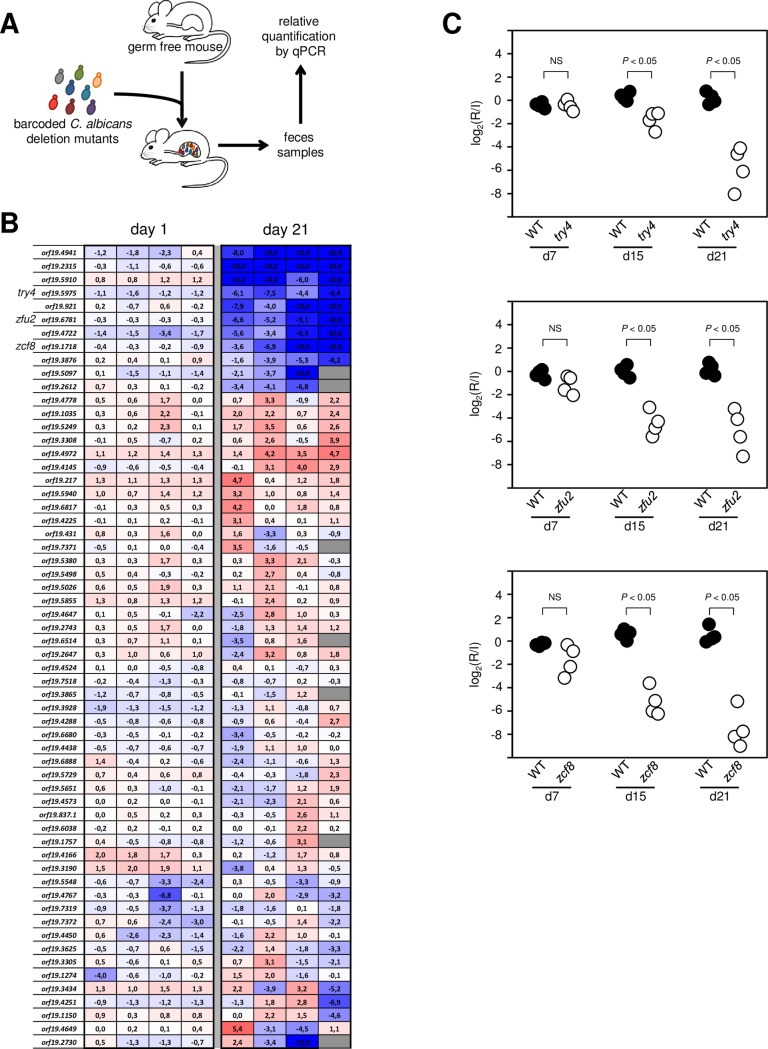
Fig 2. Identification of C. albicans transcription regulators that govern gut colonization in gnotobiotic mice.
(A) Schematic representation of the screening approach to identify genetic determinants of in vivo fitness in the fungus C. albicans. Each one of four pools consisting of 15–18 signature-tagged strains was gavaged in four germ-free mice. The abundance of each strain in the inoculum and after recovery from fecal pellets was determined by qPCR with oligos complementary to the signature tags. (B) Log2 (recovered/inoculum) values for each deletion mutant at one or 21 days post gavage (each column represents one mouse). Color intensity indicates reduction (blue) or accumulation (red). The order in which the mutants are displayed reflect hierarchical clustering. The eight mutants that cluster at the top display a consistent pattern of reduction in abundance in all mice at day 21. Four of these deletion mutants (orf19.4941, orf19.2315, orf19.921 and orf19.4722) had previously been identified in a screening conducted in the antibiotic-promoted gut colonization murine model. The deletion mutants try4Δ/Δ [orf19.5975], zfu2Δ/Δ [orf19.6781], zcf8Δ/Δ [orf19.1718], and orf19.5910Δ/Δ cluster together with the previous group but had not been identified before in any other mouse setting. (C) Validation of the gut fitness defect phenotype of three newly identified genes. Germ free animals were gavaged with 1:1 mixtures of the wild-type reference strain and try4Δ/Δ, zfu2Δ/Δ or zcf8Δ/Δ. The abundance of each strain in the inoculum (I) and after recovery from fecal pellets (R) at day 7, 15 and 21 was determined by qPCR (strains were barcoded). The deletion mutant strains were progressively depleted from the fecal pellets relative to the wild-type reference strain (P values are indicated on top of each comparison). Each circle represents the measurement from one mouse. Statistical analysis by the Mann-Whitney test. NS, not significant.