Figure 1.
Identified Variants Are Localized within Conserved Helicase Motifs of DHX30
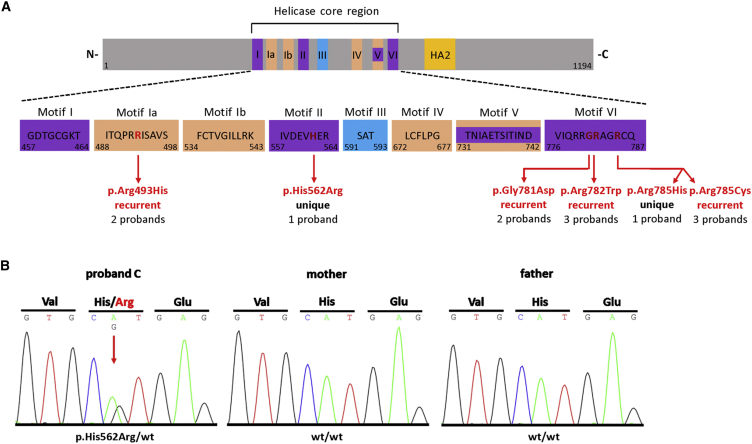
(A) Top: Schematic protein structure of DHX30 showing conserved motifs of the helicase core region and the helicase associated domain (HA2). Nucleotide-interacting motifs (I, II, and VI) are shown in purple, nucleic acid-binding motifs (Ia, Ib, and IV) in orange, motif V, which binds nucleic acid and interacts with nucleotides, in purple and orange, and motif III, which couples ATP hydrolysis to RNA unwinding, in blue. (N- N terminus; C- C terminus). Bottom: Amino acids within conserved motifs of the helicase core region. The position of the first and last amino acid within each motif is denoted below left and right, respectively. The position of the de novo mutations identified in this study are marked with vertical red arrows and shown in red.
(B) Sanger sequence electropherograms of parts of DHX30 after PCR amplification of genomic DNA of the affected individual C and his parents, exemplifying de novo status of the mutations identified here. The amino acid translation is shown in the three-letter code above the DNA sequence. The red arrow indicates the heterozygous mutation at c.1685A>G, (p.His562Arg), present only in the DNA sample of the affected individual.