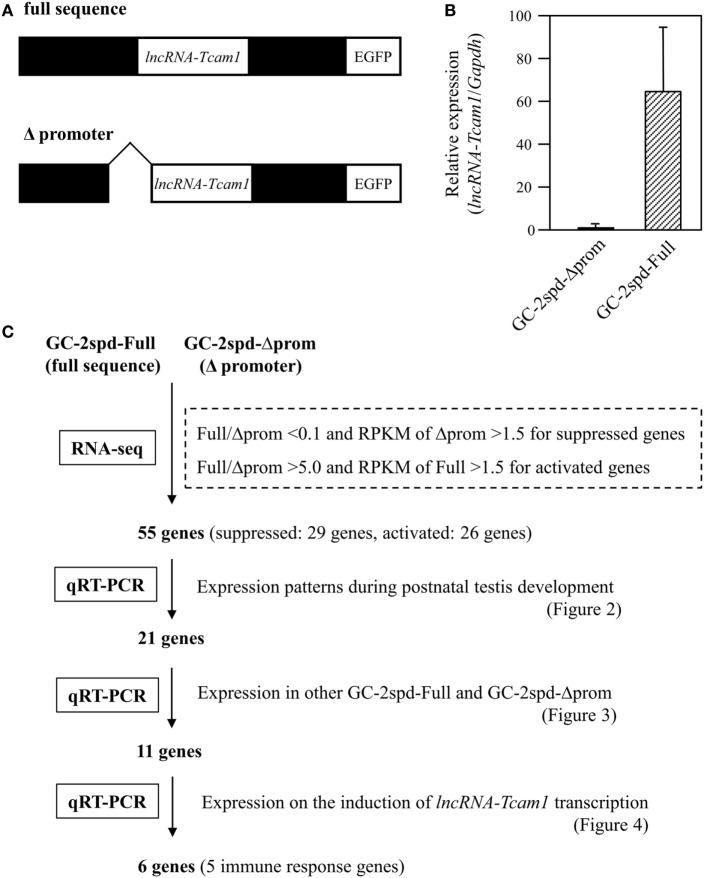
Figure 1.
A strategy to identify potential target genes of long non-coding RNA-testicular cell adhesion molecule 1 (lncRNA-Tcam1). (A) Constructs used for establishing GC-2spd(ts) cell clones that expressed lncRNA-Tcam1 at different levels. A 6.9-kb sequence encompassing the upstream and downstream regions of lncRNA-Tcam1 was obtained from a mouse BAC clone, and connected to the enhanced green fluorescent protein (EGFP) gene (full sequence). A 0.8-kb sequence corresponding to the lncRNA-Tcam1 promoter was deleted by restriction digestion (Δpromoter). The reporter EGFP gene was used to investigate the transcriptional regulation activity of the lncRNA promoter sequence in our previous study. Each construct was transfected into GC-2spd(ts) cells, and stable cell clones were established by the limited dilution method. (B) Expression of lncRNA-Tcam1 in GC-2spd-Full and GC-2spd-Δprom. GC-2spd-Δprom was derived from the cells transfected with “Δpromoter” and used as a clone expressing lncRNA-Tcam1 at a low level. GC-2spd-Full was a stably transfected clone with “full sequence” and used as the cells expressing lncRNA-Tcam1 at a high level. Total RNA was isolated from each clone, and the expression level of lncRNA-Tcam1 was measured by quantitative reverse transcription-polymerase chain reaction (qRT-PCR). The value was normalized to the Gapdh mRNA level, and the level in GC-2spd-Δprom was set to 1.0. The data are presented as mean value ± SD from three independent experiments. (C) An outline of the screening process to identify potential target genes of lncRNA-Tcam1 in this study.