Figure 3.
Analysis of the Offspring of Greb1l Mouse CRISPR-Cas9 Mutants
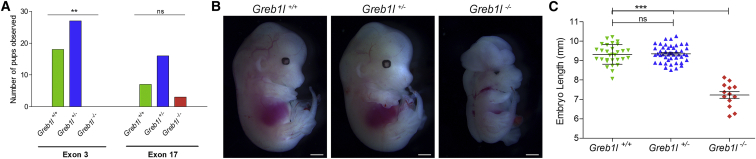
(A) Genotyping of the offspring of backcrosses and intercrosses described in Table S3B, showing embryonic lethality for Greb1l−/− embryos (homozygous or compound heterozygous frameshift mutations). Number of pups analyzed: 45 for exon 3 mutants and 26 for exon 17 mutants. The only 3 Greb1l−/− pups obtained (in red) were compound heterozygous with one of the mutations being an in-frame 3 bp deletion in exon 17 (see Table S3B). Chi-square test of observed versus expected genotypes; statistical significance ∗∗p = 0.0015.
(B) Whole-mount E13.5 embryos (delATAG mutation) showing the reduced size of Greb1l−/− embryos and the exencephaly. Scale bar, 1 mm.
(C) Quantification of the length of E13.5 embryos. Number of analyzed embryos: 30 Greb1+/+, 48 Greb1l+/−, 13 Greb1−/−. Statistical significance ∗∗∗p < 0.0001 (one-way ANOVA and Newman-Keuls multiple comparison test). Error bar: SD.