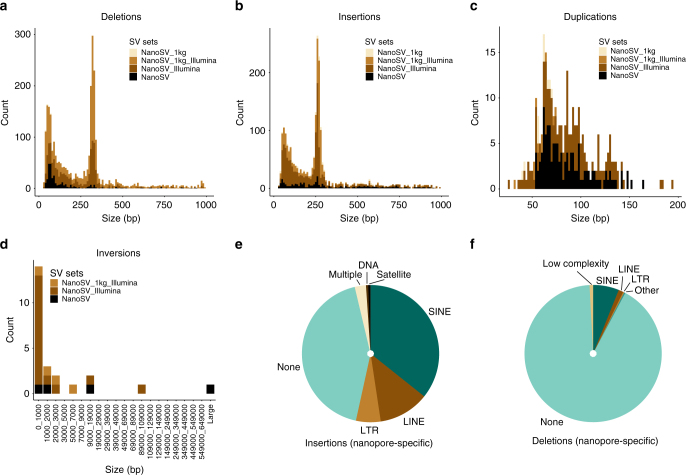
Fig. 4.
Genome-wide detection of SVs using nanopore sequencing data. a–d The total amount of high-confidence NanoSV calls for Patient 1 and Patient 2 jointly, across different SV size bins and stratified by SV type as follows: a deletions, b insertions, c duplications, and d inversions. The NanoSV calls were intersected with SV calls from other data sources (Illumina data of Patient 1 and Patient 2 and 1KG phase 3 sites). For a, b, the x-axis was trimmed to 1000 bp for visibility and a small number of variants beyond this size are not displayed in the figure. Similarly, for c, the x-axis was limited to 200 bp. e The repeat content of nanopore-specific insertions. f The repeat content of nanopore-specific deletions. Repeat annotation was obtained from the UCSC repeat masker table (GRCh37)