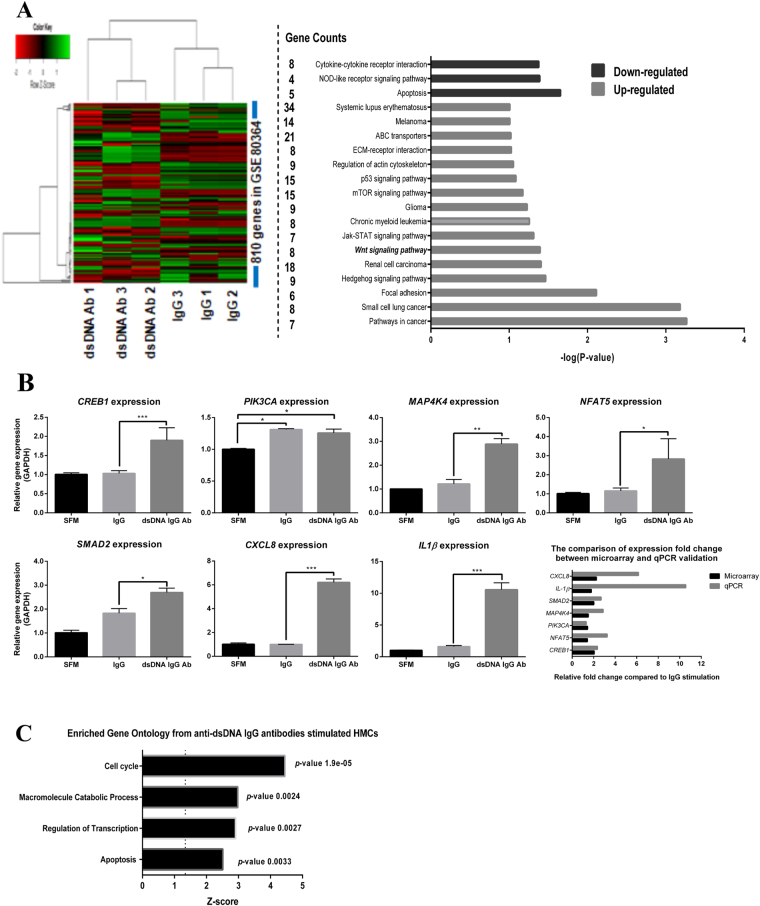
Figure 3.
Transcriptomics profiling of HMCs treated with anti-dsDNA IgG antibodies. (A) (Left) Heat-map shows the expression correlation between group of samples and significantly upregulated or downregulated genes after anti-dsDNA IgG antibody induction (fold-change > 1.2 or < 1.2). (Right) Genes that were upregulated or downregulated significantly during anti-dsDNA IgG antibody stimulation are clustered based on their involvement in the same pathway (KEGG-pathway analysis). (B) The graphs show qPCR validation from selected candidate mRNAs according to microarray result and comparison of fold change expression in microarray and qPCR. The expressions were normalized using GAPDH. (C) The graph demonstrates the microarray results using functional annotation clustering and enrichment scores with adjusted P-value in the DAVID bioinformatics resources version 6.7 (https://david.ncifcrf.gov/tools.jsp). Microarray data were analysed using lumi R-bioconductor packages. Multiple corrections (FDR) were performed by the Bonferroni-Hochberg method. The p-values < 0.05 were considered significant. qPCR data are expressed as mean ± SEM of a minimum of three biological samples from three independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001 versus scramble controls.