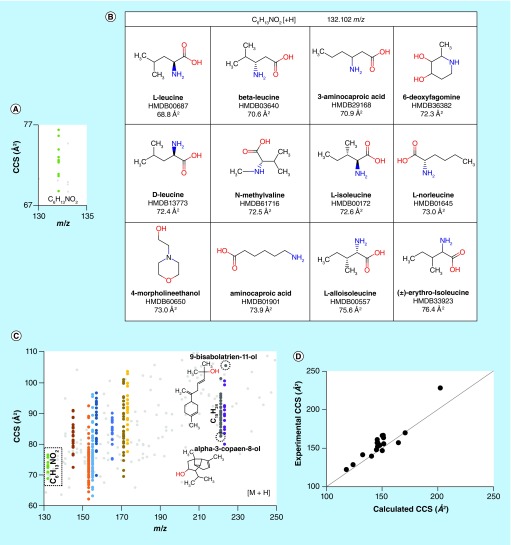
Figure 5. . Performance of the molecular modeling and quantum chemical calculation pipeline.
(A) A plot of calculated collision cross section (CCS) versus m/z for the protonated forms of the molecules shown in (B), illustrating the degree of orthogonality of the two metrics; (B) metabolite structures from the Human Metabolome Database corresponding to the formula C6H13NO2; (C) a plot of CCS versus m/z for 200 metabolites, out of the >11 k solved structures obtained from the Human Metabolome Database. Different colors correspond to CCSs for different metabolites with identical molecular formulae. The data for C6H13NO2 from (B) are outlined in the dashed box, and two anthropogenic chemicals with the formula C15H24O, 9-bisabolatrien-11-ol and α-3-copaen-8-ol are shown to have very different CCS; and (D) the theoretical CCS values obtained for selected +H, -H and +Na forms of 11 small molecules show good agreement with experimental CCS values derived from an SPE-ion mobility spectrometry (IMS)-MS platform, with average error of appproximately 2%. All CCS values were predicted using He as the buffer gas.