Fig. 3.
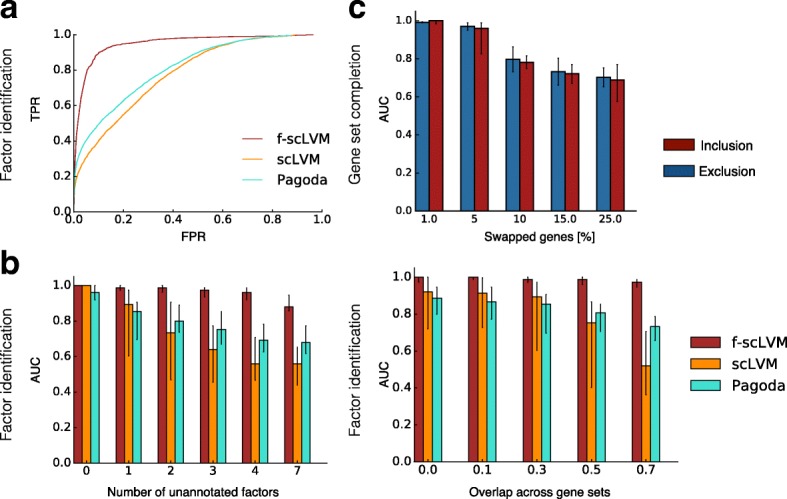
Model validation using simulated data. a, b Accuracy of f-scLVM and alternative methods for recovering the set of simulated drivers of gene expression heterogeneity. a Receiver operating characteristics (ROC) pooled across different simulated datasets (Additional file 2: Table S1). b Area under the ROC curve (AUC) when simulating an increasing number of unannotated factors not included in the pathway database (left) and when considering increasing overlap between simulated gene sets (right). c Ability of f-scLVM to augment gene sets when an increasing proportion of genes in the annotation were falsely assigned. Shown is an AUC for correctly including genes omitted from gene sets (red) and for removing genes that were incorrectly annotated (blue). Bar plots in b, c show the median AUC across 50 repeat experiments per setting with error bars corresponding to 25 and 75% percentiles. FPR false positive rate, TPR true positive rate
