Figure 1. Data-driven candidate discovery and targeted functional screen identifies putative regulators of cytosolic nucleic acid sensing in germ cells.
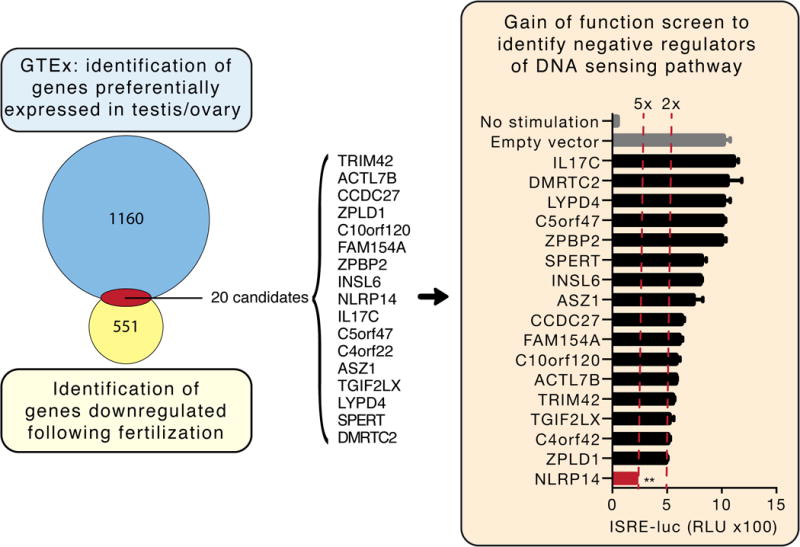
Left panel: Venn diagram of genes preferentially expressed in testis and/or ovary and down-regulated following oocyte fertilization (see Materials for further description of the discovery method). Of the 20 candidates identified, 17 (listed) were successfully cloned and expressed in 293T cells. Right panel: 293T cells stably expressing STING-HA (293T-STING-HA) were cotransfected with cGAS-Flag and each candidate along with the luciferase gene under control of the ISRE promoter and monitored for luciferase expression 24h post transfection. Shown are relative luciferase units (RLU) normalized to Renilla luciferase expression. Dotted red lines indicate 2- and 5-fold suppression. **P<0.05, Student’s test. Error bars indicate the SD. Data is representative of at least two independent experiments.
