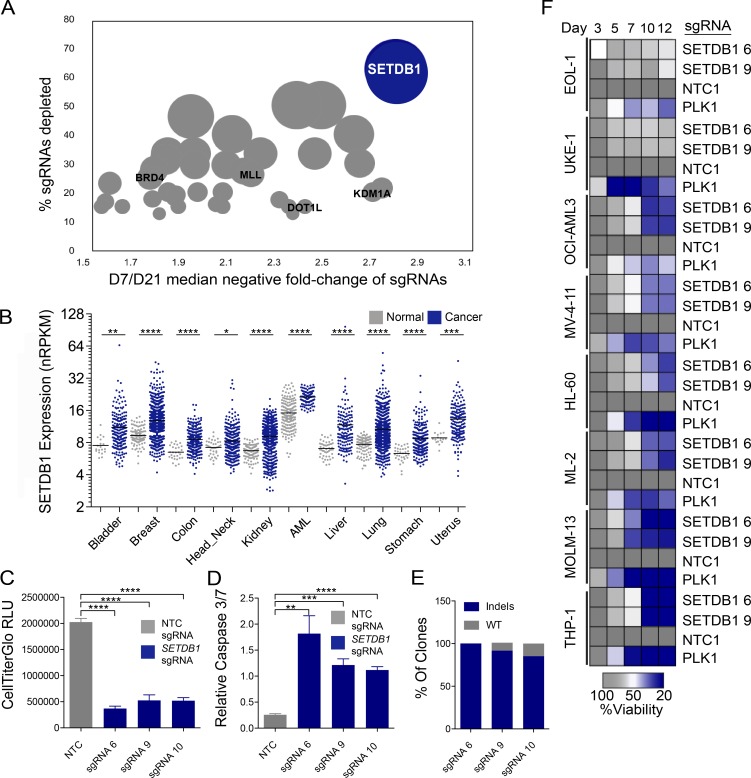
Figure 1.
A CRISPR/Cas9 genetic screen identifies SETDB1 as a critical regulator of the leukemic cell survival that is overexpressed in many cancers. (A) Median negative fold changes of sgRNA abundance at day 21 versus day 7, ranked by percentage of sgRNAs depleted for each gene in THP-1–Cas9 cells. n = 3 biological replicates for the day 7 reference and n = 2 biological replicates for day 21. The size of each circle corresponds to the fraction of depleted sgRNAs/target. Select regulators of AML cell growth are highlighted in black text. Significant fold changes were calculated with DESeq2. (B) Compiled data from TCGA RNA-seq. SETDB1-normalized RPKM (reads per kilobase of transcript per million mapped reads) values in cancer and normal tissues. Student’s t tests were performed. (C and D) Viability and Caspase 3/7 levels, as measured by Caspase-Glo, in THP-1–Cas9 cells after 7 d of treatment with three different SETDB1-specific sgRNAs or an NTC sgRNA. Mean relative light units (RLU) are shown. n = 3 experiments. Error bars represent standard deviation. Student’s t tests were performed. (B–D) *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P ≤ 0.0001. (E) Indel frequency at listed sgRNA target sites. Genomic DNA derived from THP-1–Cas9 cells 7 d after sgRNA infection. (F) Arrayed target validation screen in eight Cas9-stable AML lines. Mean viability at each day is shown. Screen performed in duplicate.