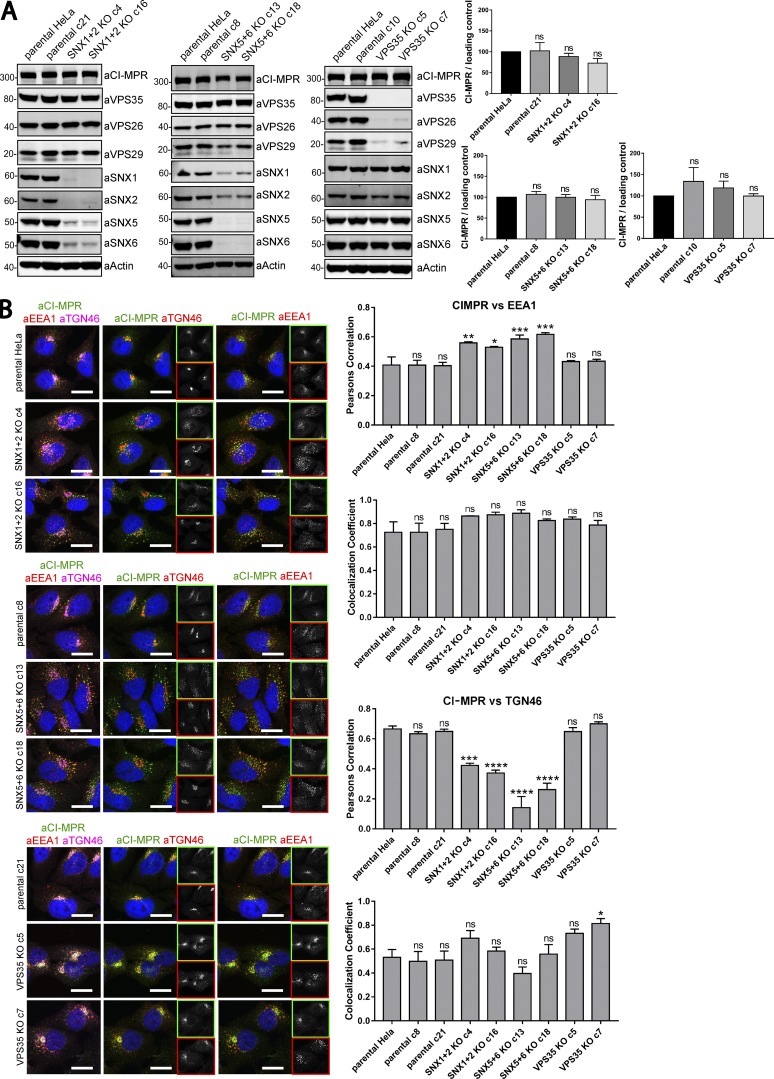
Figure 8.
Clonal HeLa KO cell lines recapitulate the SNX1/2–SNX5/6 mediated retromer-independent retrograde transport of CI-MPR. (A) Clonal cell lines were isolated from a heterogeneous population of CRISPR-Cas9 KO. Two independent lines were biochemically characterized as SNX1+2 KOs, two as SNX5+6 KOs, two as VPS35 KOs, and three as parental clonal lines. Molecular masses are given in kilodaltons. (A, right) CI-MPR levels were analyzed by Western blotting. n = 3 independent experiments. (B, left) Immunofluorescence and colocalization analysis of endogenous CI-MPR, TGN marker TGN46, and early endosomal marker EEA1 in parental HeLa and clonally selected KO lines. Bars, 20 µm. (B, top right two graphs) n = 3 independent experiments; parental HeLa, 58 cells; parental c8, 79 cells; parental c21, 81 cells; SNX1+2 KO c4, 75 cells; SNX1+2 KO c16, 70 cells; SNX5+6 KO c13, 82 cells; SNX5+6 KO c18, 89 cells; VPS35 KO c5, 83 cells; VPS35 KO c7, 72 cells. (B, bottom right two graphs) n = 3 independent experiments; parental HeLa, 67 cells; parental c8, 72 cells; parental c21, 87 cells; SNX1+2 KO c4, 83 cells; SNX1+2 KO c16, 80 cells; SNX5+6 KO c13, 84 cells; SNX5+6 KO c18, 98 cells; VPS35 KO c5, 73 cells; VPS35 KO c7, 68 cells (means ± SEM; one-way ANOVA compared with parental HeLa. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.001).