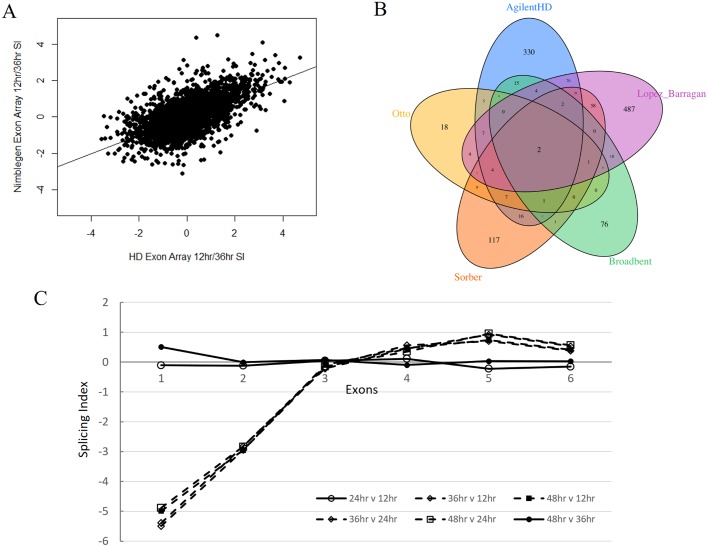
Fig 4. Exon arrays can detect exon skipping transcript isoforms.
Comparison of genes with transcript isoforms (A) Differential exon splicing is detected on both Nimblegen and Agilent HD exon array platforms using splicing index with a correlation of 0.626. (B) Venn Diagram showing the overlap of genes with transcript isoforms reported by the Agilent HD Exon Array (blue) and RNA-seq studies (Otto (yellow), Sorber (orange), Broadbent (green), and Lopez-Barragen (purple) [24,25,36,37]), (C) Alternative start sites for PF3D7_0929200 produce time point dependent exon skipping of exons 1 and 2 during the schizont parasite stage at 36 and 48 hpi.