Fig. 2.
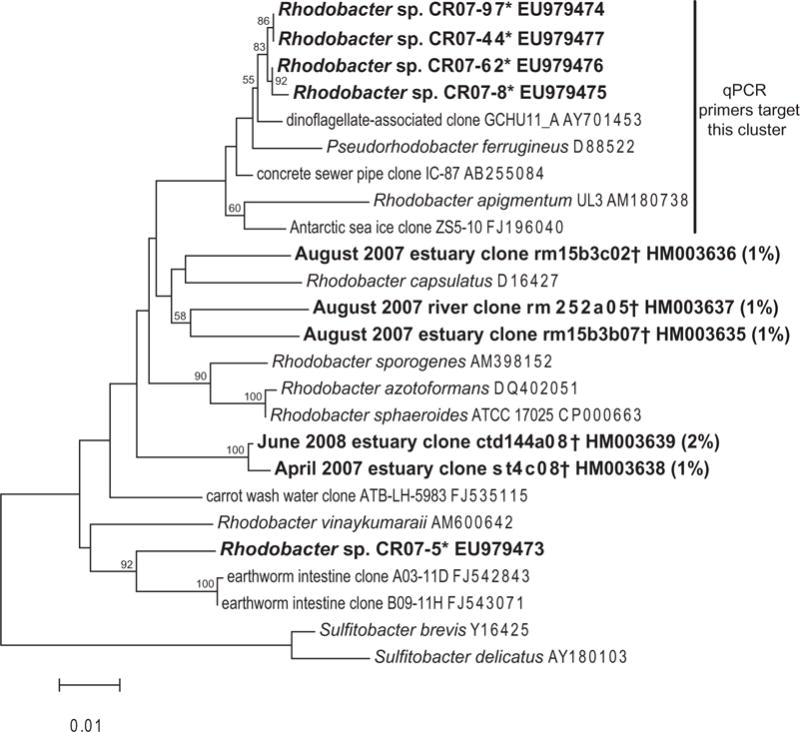
Neighbour-joining tree inferring the phylogenetic relationship between cultured strains* (asterisks) and those found in clone libraries† (daggers) in the Columbia River in this study. Alignments were created using the on-line SILVA aligner and then exported from ARB. Dendrogram was created using PHYLIP. The percentage of the sequences from each library that represented a particular OTU is given in parentheses: actual sequence abundance ranged from 1/150 to 2/80. Bootstrapping values are shown for nodes that were supported >50% of the time. Nodes with bootstrapping values >60 were also supported with maximum-likelihood analysis (data not shown). Sulfitobacter species were used as the outgroup. The cluster of Rhodobacter spp. targeted by the qPCR primers is marked by a bar.
