Figure 2.
Specificity Test of Selected sgRNAs Using T7EI Assay and NGS Analysis
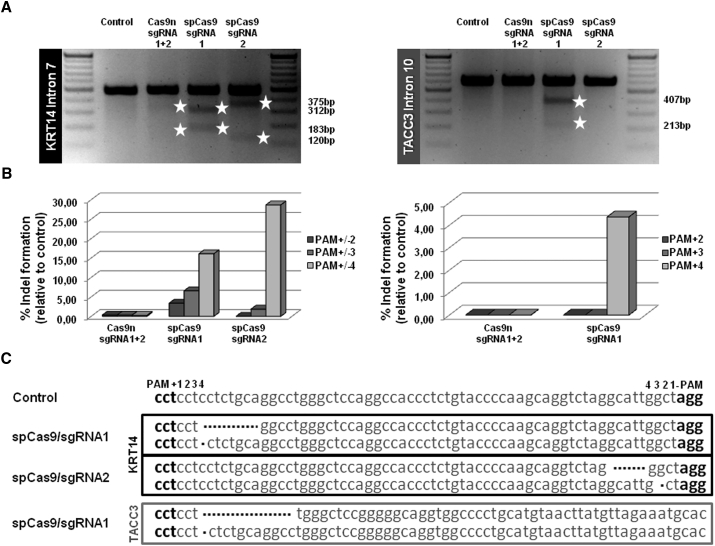
(A) Upon delivery of the respective spCas9/sgRNA (sgRNA1 and sgRNA2) expression plasmids into HEK293 cells, the target region was PCR-amplified using a primer pair binding specifically to KRT14 exon 6 and intron 7. The resulting PCR products were cleaved by T7EI treatment into two fragments, 312 and 183 bp for sgRNA1 and 375 and 120 bp for sgRNA2, thus confirming the on-target functionality of both sgRNAs. In the double-nicking approach, both sgRNAs were transfected in combination with Cas9n, resulting in the desired T7EI digestion pattern detectable via agarose gel analysis. The resulting digest pattern is a mixed pattern derived from both sgRNAs (fragments between 375 and 312 bp, and fragments between 183 and 120 bp). The predicted off-target region for sgRNA1 was PCR-amplified using a primer pair binding specifically within TACC3 intron 10. The resulting PCR products were cleaved by T7EI treatment into two fragments of 407 and 213 bp confirming the predicted off-target activity of sgRNA1 when combined with the wild-type spCas9. (B) Relative cutting efficiencies of different Cas9 enzymes were determined by next generation sequencing. Using a custom-designed panel for on/off-target analysis of the KRT14 target region and predicted off-target regions of the selected sgRNAs, the relative amount of indel formation at intron 7 of KRT14 (left panel) and intron 10 of TACC3 (right panel) was analyzed. The graphs show the relative amount of indels compared with untreated control. (C) Sequences of NGS analysis showing mainly small deletions of 1–20 bp for each spCas9/sgRNA at the KRT14 and TACC3 locus.