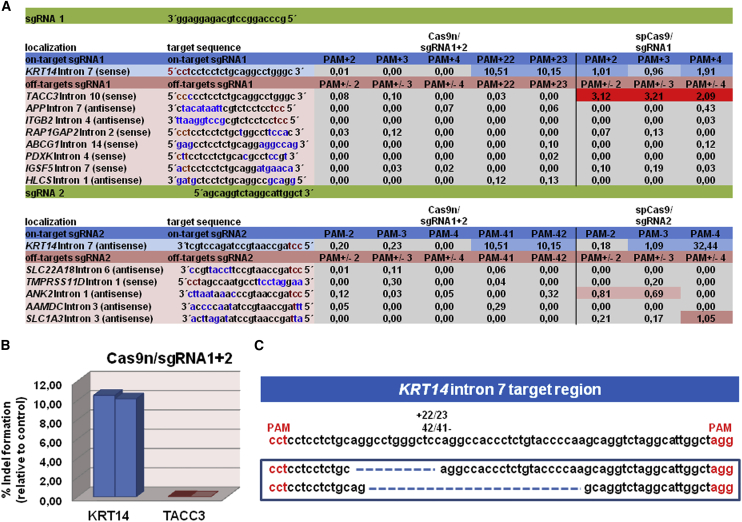
Figure 7.
Specificity Test of Selected sgRNAs in an MC-Treated EBS hKc Cell Line Using NGS
(A) DNA cleavage activities of sgRNAs at on-target and off-target regions are represented as % indels relative to untreated control. The cutoff for cleavage activity was set to 0.5%. Colored boxes indicate cleavage activity of the respective Cas9/sgRNAs at on-target and off-target regions. On-target and off-target region sequences are listed, with red letters indicating the PAM position, blue letters indicating mismatches between the sgRNA on-target binding sites and the potential off-target regions, and black letters indicating matches between on-target and off-target regions. (B) Graphs show the relative amount of indels (PAM +22/23) within KRT14 intron 7 and TACC3 intron 10, caused by double-nicking compared with untreated control. (C) Sequences of NGS analysis showing frequent deletions for the Cas9n/sgRNA1 and Cas9n/sgRNA2 configuration.