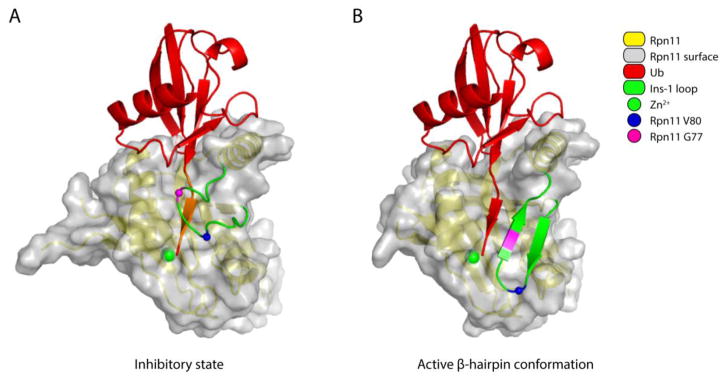
Figure 3. Conformational changes in the Ins-1 loop activate Rpn11.
(A) The inactive form of Rpn11 is based on the free Rpn11-Rpn8 model (PDB: 4O8X [64]). Ubiquitin was positioned by superimposing the ubiquitin-bound Rpn11 (PDB: 5U4P [34]) onto the free Rpn11 model. The residues in the C-terminal tail of ubiquitin that are within 1.4 Å of the Ins-1 loop are painted orange to indicate steric hindrance. (B) The active conformation of Rpn11 is based on the structure of Ub-Rpn11-Rpn8 (PDB: 5U4P [34]). In both A and B, the surface of the Ins-1 loop was removed to allow better visualization of the beta-sheet formed between the C-terminal tail of ubiquitin and this loop. Other color coding is given in the key.