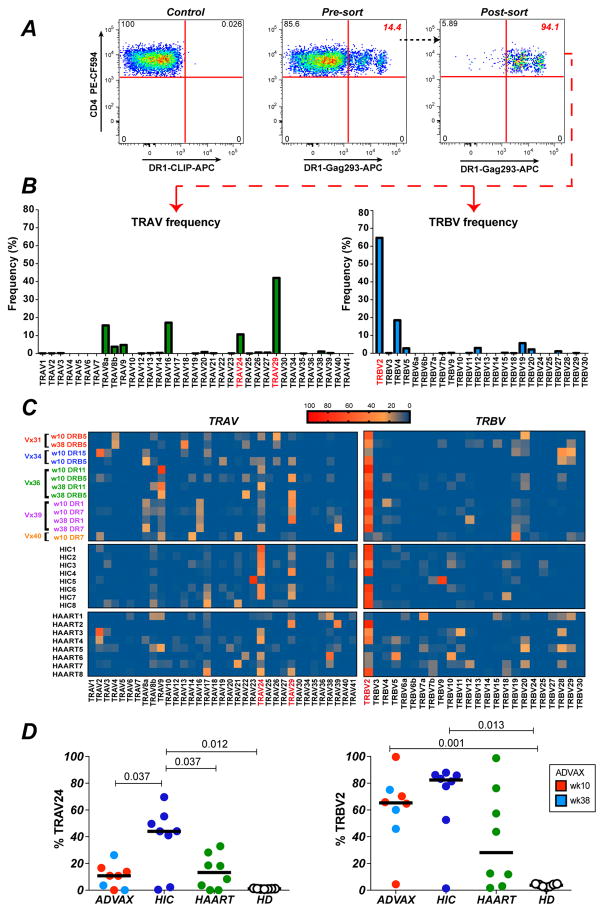
Figure 2. Biased TCR variable gene usage in Gag293-specific CD4+ T cells amplified by EP DNA vaccination.
(A) Example of Gag293-specific CD4+ T cells sorting after HLA-DR tetramer labeling. A primary CD4+ T cell line derived from an ADVAX recipient was labelled with an HLA-DRB1*0101 (DR1) tetramer loaded with Gag293. The percentage of tetramer+ (Tet+) cells in the total CD4+ T cell population (middle plot) and in the sorted Tet+ population (right plot) is reported in red. A sample labeled with the DR1 tetramer loaded with an irrelevant peptide (CLIP) was used as a negative control (left plot).
(B) Representative example of qPCR analysis (1 out of 8 experiments) to quantify the relative expression of the TRAV and TRBV variable genes in sorted Gag293-specific CD4+ T cells from an ADVAX recipient.
(C) Heatmaps depicting the TRAV and TRBV gene usage in Gag293-specific CD4+ T cells from vaccinee samples (n=5) sorted with different HLA-DR tetramers. The TRAV and TRBV gene usage in Gag293-specific CD4+ T cells from HIV controllers (HIC) and treated patients (HAART) are reported below. The most prevalent TRAV and TRBV genes are marked in red.
(D) Comparison of the expression of TRAV24 and TRBV2 in Gag93-specific cells from vaccinees (week 10, red, n=5; week 38, light blue, n=3), HIV controllers (HIC, dark blue, n=8) and treated patients (HAART, green, n=8). The frequency of TRAV24 and TRBV2 in the total TCR repertoire from healthy donors (HD, open circles, n=7) is reported for comparison. Significant differences (P < 0.05) obtained with the Mann-Whitney U test are reported.