FIG 1 .
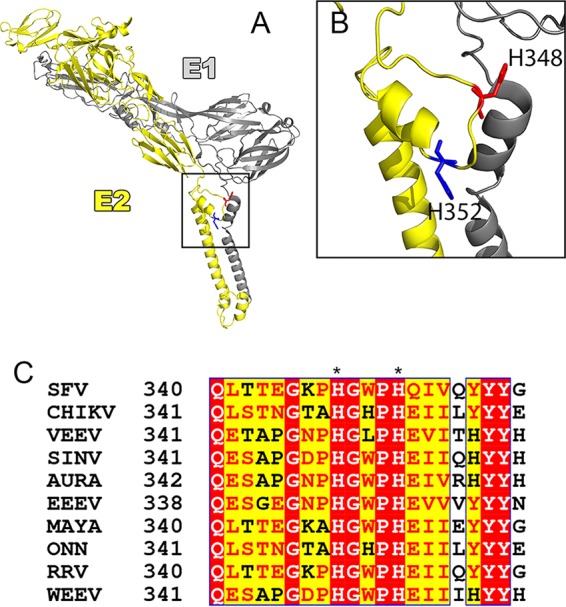
The E2 D-loop and the highly conserved H348 and H352 histidines. (A) Model of the VEEV surface glycoproteins showing E1 in gray and E2 in yellow, with H348 shown in red and H352 in blue (residues are numbered as for SFV [VEEV, H349 and H53]) (PDB accession no. 3J0C). (B) Expanded view of the boxed area in panel A showing the E2 D-loop with H348 and H352. The images in panels A and B were prepared using PyMOL software (the PyMOL Molecular Graphics System, v1.2r2; Schrödinger, LLC). (C) Alignment of E2 proteins from members of the Alphavirus genus, demonstrating that H348 and H352 are highly conserved (numbering as described for SFV). Red highlighting indicates high sequence conservation, and yellow highlighting indicates some sequence conservation, with conserved residues shown in red text. Alignment was performed using NPS@ (58) with CLUSTAL W (59). Alignment formatting was carried out using ESPript (60). SINV, Sindbis virus; AURA, Aura virus; EEEV, Eastern equine encephalitis virus; MAYA, Mayaro virus; ONN, O'nyong-nyong virus; RRV, Ross River virus; WEEV, Western equine encephalitis virus.
