Abstract
Background
Non small cell lung cancer (NSCLC) is diagnosed in most cases on small tissue samples, such as cytological preparations and histological biopsies; these limited tissue specimens may be not always sufficient for testing epidermal growth factor receptor (EGFR) mutations and other relevant predictive biomarkers. Cell-free DNA (cfDNA) can be used as a surrogate for EGFR mutational testing, whenever tissue is unavailable. However, the detection of gene mutations on cfDNA is challenging; in fact, the extremely low concentration of circulating tumor DNA requires the implementation of highly sensitive and validated next generation techniques.
Methods
Thus, we have recently validated a novel next generation sequencing (NGS) assay, employing the SiRe® gene panel to detect on cfDNA mutations of EGFR and KRAS, NRAS, BRAF, cKIT and PDGFR genes. In this current study, we report on a series of NSCLC patients, without available tissue for EGFR testing, who prospectively underwent SiRe® NGS analysis.
Results
The results confirm the high clinical performance, in terms of success rate and mutation detection, of NGS based analysis of cfDNA.
Conclusions
SiRe® NGS panel represent an effective diagnostic tool in cfDNA analysis setting.
Keywords: Liquid biopsy, non small cell lung cancer (NSCLC), next generation sequencing (NGS), tyrosine kinase inhibitors (TKIs)
Introduction
Non small cell lung cancer (NSCLC) is diagnosed in most cases at advanced stages of disease. Diagnostic samples are frequently scarcely cellular, being represented by either cytological specimens or small tissue endoscopic biopsies; these limited tissue samples often may be not sufficient for epidermal growth factor receptor (EGFR) and other clinical relevant biomarkers, such as ALK translocation and PD-L1 expression, whose assessment is required to select patients for first line treatment administration (1,2). In particular for EGFR tyrosine kinase inhibitors (TKIs), such as gefitinib, erlotinib and afatinib the identification of activating EGFR mutations in exon 18, 19 and 21 is mandatory before the first line treatment (3-8). To date according to the European Medicines Agency guidelines, in patients without tissue availability, only for EGFR TKIs treatment decision making, cell-free DNA (cfDNA) can be used as a fast and non-invasive surrogate for EGFR mutational testing (9-13). However, the assessment of gene mutations in cfDNA is challenging, in particular in basal setting, for the detection of first and second TKIs generation EGFR sensitizing mutations, due to the very low concentration of circulating tumor DNA, that represent only a small fraction of the total cfDNA (9,10,12-15). Thus, the clinical implementation of next generation techniques, such as next generation sequencing (NGS) or digital PCR (dPCR) based assay is crucial (9,10,12,13,16,17). In a recent study of ours, we validated the SiRe® NGS panel for mutation detection in EGFR, KRAS, NRAS, BRAF, cKIT and PDGFR starting from cfDNA retrieved from patients with different solid tumors (NSCLC, metastatic colo-rectal cancer, melanoma and gastrointestinal stromal tumor) (13). SiRe®, with a lower limit of detection of 0.01% and a reference range of 568 clinical relevant mutations, showed a higher analytical performance respect to a very sensitive modified TaqMan probe real time PCR based approach (13). In the clinical trials settings and in other published validation studies (18-20), the analysis of cfDNA gene mutations was carried out using as gold standard the mutational status obtained on matched tissue derived DNA, but little is known regarding the application of this approach in clinical setting, in particular in baseline setting of NSCLC patients, prior to EGFR TKIs administration, without a referent DNA derived tissue to confirm the mutational data obtained on cfDNA (21).
In this current study, we reviewed the NGS data obtained by using SiRe® NGS panel starting from cfDNA collected in routine NSCLC baseline setting to prospectively select patients, without tissue availability, for first and second generation EGFR TKIs treatment administration.
Methods
Our molecular laboratory is an accredited Italian reference center for predictive molecular pathology testing in oncology (22). In particular for NSCLC patients, from January 2017 to March 2017, n=64 liquid biopsy analysis was requested from the oncologists of different South Italy institutions (n=14), following the European Medicines Agency guidelines, for the analysis of EGFR mutations on cfDNA inpatients without tissues availability at presentation, to assess the eligibility to first and second generation EGFR TKIs (Table 1). On the overall n=39 men and n=25 women were analyzed with a mean age of 66 years (range, 36–89 years). For each patient, 10 mL of blood was collected in-house by using EDTA Vacutainer tubes (BD, Plymount, UK) by a dedicated nurse at the Department of Public Health of the University of Naples Federico II. The protocols adopted in this study were previously validated (13). Briefly, before cfDNA extraction, two centrifugation steps (2,300 rpm for 10 min) were carried out to obtain at least 1.2 mL of plasma for each patient. cfDNA was extracted by using the QIAsymphony DSPVirus/Pathogen Midi Kit on the QIAsymphony robot (Qiagen, Venlo Limburg) accordingly with the manufacturer instructions. By using SiRe® panel, following the previously validated protocol, libraries were automated constructed and purified using Ion AmpliSeq DL8 Kit (Thermofisher) on the Ion Chef instrument (Thermofisher) and, after barcoding, purified libraries derived from eight cfDNA plasma samples were diluted and combined with eight additional cfDNA-derived libraries to obtain a 16 Ion Code pooled library, re-loaded into the Ion Chef instrument for template preparation by using the Ion PGM Hi-Q IC Kit (Thermofisher). Finally, templates were loaded into the 316v2 chip and sequenced on Personal Genome Machine (PGM). Signal processing and base calling were carried out using the default base-caller parameters on Torrent Suite (v.5.0.2) and coverage analysis was performed using SiRe® specific bed files with coverage plug-in (v.5.0.2.0). In addition to automatic variant calling analysis, by using SiRe® panel specific optimized variant caller plug-in (v.5.0.2.1) parameters, BAM files were visually inspected with the Golden Helix Genome Browser v.2.0.7 (Bozeman, MT, USA). Only variants with >5× allele coverage and a quality score >20, within an amplicon coverage at least 1,000× alleles, were reported and the relative mutated allele frequency was annotated, considering not only EGFR, but also KRAS, BRAF and NRAS gene hot-spots region, relevant for NSCLC and covered by the SiRe® panel.
Table 1. Patients characteristics and NGS results.
| Patient | Sex | Age | Reads | Mean read length (bp) | Number of mapped reads | % reads on target (%) | Average reads per amplicon | Uniformity of amplicon coverage (%) | NRAS (allelic frequency) | BRAF (allelic frequency) | EGFR (allelic frequency) | KRAS (allelic frequency) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 76 | 325,098 | 127 | 324,684 | 97.31 | 7,523 | 100.00 | WT | WT | WT | WT |
| 2 | F | 77 | 223,871 | 130 | 223,209 | 97.71 | 5,187 | 100.00 | WT | WT | WT | WT |
| 3 | M | 68 | 288,543 | 127 | 287,410 | 97.60 | 6,679 | 100.00 | WT | WT | WT | WT |
| 4 | F | 74 | 26,017 | 125 | 25,666 | 96.70 | 590.9 | 97.62 | G12S (1.00%) | WT | WT | WT |
| 5 | F | 36 | 163,218 | 128 | 162,752 | 98.06 | 3,800 | 97.62 | WT | WT | WT | G12D (1.50%) |
| 6 | F | 50 | 146,656 | 127 | 145,906 | 97.25 | 3,378 | 100.00 | WT | V600A (0.20%) | WT | WT |
| 7 | M | 70 | 173,021 | 127 | 172,314 | 97.55 | 4,002 | 97.62 | G13D (0.34%) | WT | WT | Q61H (0.20%) |
| 8 | M | 64 | 151,445 | 127 | 150,370 | 97.03 | 3,474 | 97.62 | WT | WT | WT | G12C (1.30%) |
| 9 | M | 61 | 75,353 | 127 | 74,972 | 97.68 | 1,744 | 97.62 | WT | WT | WT | WT |
| 10 | M | 45 | 190,620 | 126 | 189,708 | 97.49 | 4,403 | 100.00 | WT | WT | H773_V774insH (37.0%) | WT |
| 11 | M | 48 | 103,023 | 128 | 102,573 | 97.79 | 2,388 | 97.62 | WT | WT | WT | G12C (0.60%) |
| 12 | F | 61 | 163,363 | 128 | 162,841 | 97.75 | 3,790 | 97.62 | WT | WT | WT | WT |
| 13 | F | 57 | 130,596 | 128 | 130,250 | 97.72 | 3,031 | 97.62 | WT | WT | WT | WT |
| 14 | F | 51 | 155,551 | 128 | 155,018 | 97.40 | 3,595 | 97.62 | WT | WT | WT | WT |
| 15 | M | 62 | 60,927 | 128 | 60,611 | 97.77 | 1,411 | 97.62 | WT | WT | V769_D770insASV (12.30%) | WT |
| 16 | F | 87 | 67,571 | 128 | 67,404 | 97.85 | 1,570 | 97.62 | WT | WT | WT | WT |
| 17 | F | 77 | 181,295 | 129 | 180,633 | 98.06 | 4,217 | 95.24 | WT | WT | WT | WT |
| 18 | M | 75 | 104,653 | 127 | 104,925 | 97.79 | 2,429 | 97.62 | WT | WT | WT | WT |
| 19 | M | 58 | 262,771 | 130 | 261,202 | 97.94 | 6,091 | 97.62 | WT | WT | WT | WT |
| 20 | F | 38 | 248,897 | 128 | 248,287 | 97.68 | 5,775 | 97.62 | WT | WT | WT | WT |
| 21 | F | 51 | 169,989 | 128 | 169,581 | 97.44 | 9,734 | 97.62 | WT | WT | WT | WT |
| 22 | M | 64 | 36,774 | 128 | 36,693 | 97.84 | 854.7 | 97.62 | WT | WT | WT | WT |
| 23 | M | 74 | 195,245 | 128 | 194,624 | 97.50 | 4,518 | 100.00 | WT | WT | L858R (3.20%) | WT |
| 24 | F | 54 | 82,480 | 128 | 82,303 | 97.80 | 1,916 | 97.62 | WT | WT | WT | WT |
| 25 | M | 84 | 12,499 | 127 | 12,439 | 97.79 | 289.6 | 97.62 | WT | WT | WT | WT |
| 26 | F | 59 | 71,556 | 129 | 71,360 | 97.54 | 1,657 | 97.62 | WT | WT | WT | G12D (1.30%) |
| 27 | M | 68 | 62,909 | 128 | 62,723 | 97.61 | 1,458 | 97.62 | WT | WT | WT | WT |
| 28 | M | 56 | 51,993 | 128 | 51,453 | 97.44 | 1,194 | 100.00 | WT | WT | WT | WT |
| 29 | M | 89 | 21,306 | 127 | 21,126 | 97.12 | 488.5 | 97.62 | WT | WT | WT | G12D |
| 30 | M | 53 | 76 | 60 | 66 | 39.39 | 0.69 | 91.45 | Failed | Failed | Failed | Failed |
| 31 | F | 67 | 49,539 | 129 | 49,435 | 97.50 | 1,148 | 97.62 | WT | G469A (5.00%) | WT | WT |
| 32 | M | 62 | 113,202 | 130 | 112,930 | 98.01 | 2,635 | 97.62 | WT | WT | WT | G12C (5.63%) |
| 33 | M | 70 | 72,936 | 129 | 72,752 | 97.77 | 1,694 | 97.62 | WT | WT | WT | WT |
| 34 | F | 71 | 133,127 | 129 | 132,803 | 97.93 | 3,096 | 97.62 | WT | WT | WT | WT |
| 35 | F | 75 | 78,508 | 128 | 78,340 | 97.55 | 1,820 | 97.62 | WT | WT | WT | WT |
| 36 | M | 80 | 175,862 | 130 | 175,177 | 97.66 | 4,073 | 97.62 | WT | WT | WT | WT |
| 37 | M | 66 | 118,804 | 129 | 118,575 | 97.87 | 2,763 | 97.62 | WT | WT | WT | WT |
| 38 | F | 71 | 139,472 | 129 | 139,008 | 97.23 | 3,218 | 100.00 | WT | WT | WT | WT |
| 39 | M | 67 | 111,500 | 128 | 110,909 | 97.27 | 2,571 | 97.62 | WT | WT | WT | WT |
| 40 | M | 64 | 348,181 | 135 | 347,148 | 97.91 | 8,093 | 97.62 | WT | WT | WT | G13S (0.20%) |
| 41 | M | 80 | 131,590 | 129 | 131,257 | 97.98 | 3,062 | 97.62 | WT | WT | WT | WT |
| 42 | M | 80 | 141,018 | 129 | 140,564 | 98.15 | 3,285 | 97.62 | Q61P (0.14%) | WT | WT | WT |
| 43 | F | 76 | 72,635 | 128 | 72,364 | 97.77 | 1,685 | 97.62 | WT | WT | WT | G13D (0.30%) |
| 44 | M | 42 | 152,707 | 127 | 152,031 | 97.58 | 3,532 | 97.62 | A59C (0.20%) | WT | WT | WT |
| 45 | M | 66 | 74,945 | 127 | 74,630 | 97.64 | 1,735 | 97.62 | WT | WT | WT | WT |
| 46 | F | 57 | 50,945 | 128 | 50,607 | 97.58 | 1,176 | 97.62 | WT | WT | WT | WT |
| 47 | M | 67 | 84,001 | 130 | 83,802 | 97.69 | 1,949 | 97.62 | WT | WT | WT | WT |
| 48 | M | 69 | 133,275 | 132 | 132,908 | 97.57 | 3,087 | 97.62 | WT | WT | WT | G12S (6.40%) |
| 49 | M | 65 | 92,859 | 126 | 91,962 | 96.90 | 2,122 | 97.62 | WT | WT | WT | G12C (3.30%) |
| 50 | M | 77 | 47,845 | 130 | 47,653 | 97.32 | 1,104 | 97.62 | WT | WT | WT | Q61H (4.50%) |
| 51 | M | 82 | 23,283 | 129 | 23,118 | 97.44 | 5,363 | 97.62 | WT | WT | WT | WT |
| 52 | F | 77 | 31,713 | 129 | 31,599 | 97.86 | 9,363 | 97.62 | WT | WT | WT | WT |
| 53 | M | 84 | 121,037 | 129 | 120,379 | 97.44 | 2,793 | 97.62 | WT | WT | WT | G13S (0.20%) |
| 54 | F | 73 | 73,937 | 129 | 73,230 | 97.42 | 1,699 | 97.62 | WT | WT | WT | WT |
| 55 | M | 66 | 136,734 | 131 | 136,185 | 97.37 | 3,157 | 97.62 | WT | WT | WT | WT |
| 56 | M | 69 | 102,126 | 130 | 101,718 | 97.30 | 2,356 | 97.62 | WT | WT | WT | WT |
| 57 | M | 65 | 103,211 | 131 | 102,957 | 97.48 | 2,390 | 97.62 | WT | WT | WT | WT |
| 58 | M | 65 | 73,517 | 129 | 73,310 | 97.32 | 1,699 | 97.62 | WT | WT | WT | WT |
| 59 | M | 81 | 163,403 | 130 | 162,818 | 97.33 | 3,773 | 97.62 | WT | WT | WT | WT |
| 60 | M | 68 | 182,161 | 130 | 181,087 | 97.19 | 4,190 | 97.62 | WT | WT | WT | WT |
| 61 | F | 54 | 64,662 | 127 | 63,955 | 97.52 | 1,485 | 97.62 | WT | WT | WT | WT |
| 62 | F | 64 | 119,320 | 130 | 118,669 | 97.21 | 2,747 | 97.62 | WT | WT | WT | A59V (0.20%) |
| 63 | F | 67 | 103,023 | 128 | 102,573 | 97.79 | 2,388 | 97.62 | WT | WT | ELREA (5.40%) | WT |
| 64 | F | 61 | 173,021 | 127 | 172,314 | 97.55 | 4,002 | 97.62 | WT | WT | ELREA (0.70%) | WT |
Patients characteristics, SiRe® next generation sequencing (NGS) panel run metric parameters (reads, mean read length in base pair, number of mapped reads, percentage of read on target, average reads per amplicon, uniformity of amplicon coverage) and genes mutational status with relative mutated allele frequency are reported for each sample.
Written informed consent was obtained from all patients and documented in accordance with the general authorization to process personal data for scientific research purposes from ‘The Italian Data Protection Authority’ (http://www.garanteprivacy.it/web/guest/home/docweb/docwebdisplay/export/2485392) and all samples were handled in compliance with the Helsinki Declaration (http://www.wma.net/en/30publications/10policies/b3/).
Results
The clinical performance of the SiRe® panel in basal setting was assessed by prospectively testing the plasma derived cfDNA of n=64 NSCLC patients for whom no tissue was available to test the EGFR mutational status for TKIs treatment administration. The SiRe® NGS analysis results were adequate in 98.4% of cases (63/64) accordingly to the quality parameters reported in the methods section and previously validated; only one cases (#30) failed to reach the quality thresholds for data analysis. Regarding the run metrics parameters (Table 1), the median number of reads for sample was 120,960, the median number of read length was 127 bp, the median number of mapped reads was 120,498, the mean percentage of reads on target was 97%, the average reads for amplicon was 2,894 and the uniformity of coverage was 98%, in accordance with the data obtained in our previous validation study (13). On the overall, considering EGFR, KRAS, NRAS and BRAF genes, 24 patients (38%) showed at least one mutation. Only one patient (#7) showed two concomitant mutations (NRAS p.G13D and KRAS p.Q61H). In particular, 5 EGFR mutations (8%) were detected [n=2, exon 19 deletions (both p.E746_A750delELREA); n=2, exon 20 insertions (p.H773_V774insH and V769_D770insASV); and n=1, p.L858R exon 21 point mutation]; 14 KRAS point mutations (22%) [n=11, exon 2 mutations (n=4 p.G12C, n=3 p.G12D, n=1 p.G12S, n=1 p.G13D and n=2 p.G13S); and n=3, exon 3 point mutations (n=1 p.A59V and n=2 p.Q61H)]; n=4 NRAS point mutations (6%) [n=2, exon 2 mutations (n=1 p.G12S and n=1 p.G13D); and n=2, exon 3 point mutations (n=1 p.A59C and n=1 Q61P)]; 2 (3%) BRAF point mutations [n=1 exon 11 p.G469A mutation and n=1 exon 15 p.V600E mutation]. The mutated allele frequency for each mutation detected is reported in Table 1.
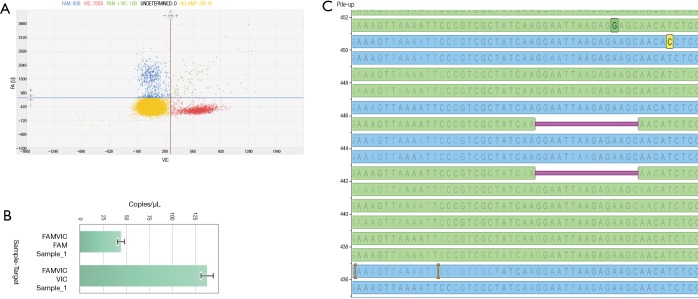
Prior to clinical reporting, only the EGFR detected mutations by the SiRe® panel were also confirmed by digital PCR based assay. An example of this approach was showed in Figure 1.
Figure 1.
Case n.64 is reported. Digital PCR Quant Studio 3D cloud software (Thermofisher) was used to analyze the scatter plot (A) and the copies of mutated and wild type alleles detected in one µl of the extracted cell-free DNA (cfDNA) (B). In the panel (C), the SiRe® panel next generation sequencing (NGS) result is reported obtained on the same extracted cfDNA and analyzed by using Golden Helix Genome Browser v.2.0.7 (Bozeman, MT, USA) and showing an epidermal growth factor receptor (EGFR) exon 19 deletion (p.E746_A750delELREA).
Discussion
Data, generated by the SiRe® NGS panel on cfDNA, prospectively collected from NSCLC patients, without tissue availability, examined for first and second generation EGFR TKIs treatment administration, are here reported; the performance of this NGS panel designed to cover only the current clinical relevant mutations, was more than excellent.
Our data confirm previous validation data. Preliminary, we had prospectively analyzed a total of 79 NSCLC patients on cfDNA. In 46 instances, cfDNA had been derived from NSCLC patients at presentation; in this subset, we detected four EGFR mutations (8.7%); more in details, these were one point mutation in exon 18 (p.G719A), two deletions in exon 19 (both p.E746_A750delELREA) and one insertion in exon 20 (p.H773-V774insH) (13). Here, in this current subsequent study, we detected two exon 19 deletions (both p.E746_A750delELREA), two exon 20 insertions (p.H773_V774insH; V769_D770insASV) and one p.L858R exon 21 point mutation. Thus, we confirm an overall EGFR mutation rate of 8.0%. In all instances, the EGFR mutations were always confirmed by an independent orthogonal dPCR based assay (Figure 1). In addition, in the present study we have also sequenced, in the same sample set, KRAS, NRAS and BRAF NSCLC relevant hot-spot regions, reporting an overall mutation rate of 38%. In particular, we detected 22% KRAS, 6% NRAS and 3% BRAF mutated samples, with only one patient that showed two concurrent mutations (NRAS p.G13D and KRAS p.Q61H). It is remarkable to note that the mutation distribution in cfDNA of this NSCLC baseline patient series was very similar to that reported on tissues derived DNA by previous studies exploiting a multi-gene assay in NSCLC (18-21).
As a general rule, in the clinical trial settings the analysis of cfDNA had as a reference the mutational status obtained on tissue derived DNA (18-20). Conversely, following the European Medicines Agency guidelines, in baseline setting, the cfDNA analysis is indicated only for those patients in which tissues is not available. For this reason, the ability of SiRe®, to detect also mutation in KRAS, NRAS and BRAF genes, offer an internal control in patients that do not show alterations in EGFR, considering that in the most part of the cases these mutations in these genes are mutually exclusive.
In conclusion, our data update and confirm that SiRe® NGS panel represents a robust analytical tool for a centralized laboratory enabling the possibility to test cfDNA mutational status in basal setting of NSCLC patients when no tissue samples are available to assess EGFR mutational status for first line treatment decision making.
Acknowledgements
This study was supported by the Department of Public Health, University of Naples Federico II.
Ethical Statement: The study was approved by Comitato Etico Università Federico II (No. 257/16) and written informed consent was obtained from all patients.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Ofiara LM, Navasakulpong A, Ezer N, et al. The importance of a satisfactory biopsy for the diagnosis of lung cancer in the era of personalized treatment. Curr Oncol 2012;19:S16-23. 10.3747/co.19.1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang S, Yu B, Ng CC, et al. The suitability of small biopsy and cytology specimens for EGFR and other mutation testing in non-small cell lung cancer. Transl Lung Cancer Res 2015;4:119-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malapelle U, Bellevicine C, De Luca C, et al. EGFR mutations detected on cytology samples by a centralized laboratory reliably predict response to gefitinib in non-small cell lung carcinoma patients. Cancer Cytopathol 2013;121:552-60. 10.1002/cncy.21322 [DOI] [PubMed] [Google Scholar]
- 4.Chan BA, Hughes BG. Targeted therapy for non-small cell lung cancer: current standards and the promise of the future. Transl Lung Cancer Res 2015;4:36-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Köhler J, Schuler M. Afatinib, erlotinib and gefitinib in the first-line therapy of EGFR mutation-positive lung adenocarcinoma: a review. Onkologie 2013;36:510-8. 10.1159/000354627 [DOI] [PubMed] [Google Scholar]
- 6.Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. 10.1016/S1470-2045(11)70393-X [DOI] [PubMed] [Google Scholar]
- 7.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. 10.1056/NEJMoa0810699 [DOI] [PubMed] [Google Scholar]
- 8.Wu YL, Zhou C, Hu CP, et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised phase 3 trial. Lancet Oncol 2014;15:213-22. 10.1016/S1470-2045(13)70604-1 [DOI] [PubMed] [Google Scholar]
- 9.Crowley E, Di Nicolantonio F, Loupakis F, et al. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol 2013;10:472-84. 10.1038/nrclinonc.2013.110 [DOI] [PubMed] [Google Scholar]
- 10.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2011;11:426-37. 10.1038/nrc3066 [DOI] [PubMed] [Google Scholar]
- 11.Karachaliou N, Mayo-de las Casas C, Queralt C, et al. Association of EGFR L858R Mutation in Circulating Free DNA With Survival in the EURTAC Trial. JAMA Oncol 2015;1:149-57. 10.1001/jamaoncol.2014.257 [DOI] [PubMed] [Google Scholar]
- 12.Malapelle U, Pisapia P, Rocco D, et al. Next generation sequencing techniques in liquid biopsy: focus on non-small cell lung cancer patients. Transl Lung Cancer Res 2016;5:505-10. 10.21037/tlcr.2016.10.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malapelle U, Mayo de-Las-Casas C, Rocco D, et al. Development of a gene panel for next-generation sequencing of clinically relevant mutations in cell-free DNA from cancer patients. Br J Cancer 2017;116:802-10. 10.1038/bjc.2017.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qin Z, Ljubimov VA, Zhou C, et al. Cell-free circulating tumor DNA in cancer. Chin J Cancer 2016;35:36. 10.1186/s40880-016-0092-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24. 10.1126/scitranslmed.3007094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bennett CW, Berchem G, Kim YJ, et al. Cell-free DNA and next-generation sequencing in the service of personalized medicine for lung cancer. Oncotarget 2016;7:71013-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vendrell JA, Mau-Them FT, Béganton B, et al. Circulating Cell Free Tumor DNA Detection as a Routine Tool forLung Cancer Patient Management. Int J Mol Sci 2017;18. pii: E264. 10.3390/ijms18020264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jänne PA, Yang JC, Kim DW, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med 2015;372:1689-99. 10.1056/NEJMoa1411817 [DOI] [PubMed] [Google Scholar]
- 19.Goss G, Tsai CM, Shepherd FA, et al. Osimertinib for pretreated EGFRThr790Met-positive advanced non-small-cell lung cancer (AURA2): a multicentre, open-label, single-arm, phase 2 study. Lancet Oncol 2016;17:1643-52. 10.1016/S1470-2045(16)30508-3 [DOI] [PubMed] [Google Scholar]
- 20.Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med 2017;376:629-40. 10.1056/NEJMoa1612674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thress KS, Brant R, Carr TH, et al. EGFR mutation detection in ctDNA from NSCLC patient plasma: A cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer 2015;90:509-15. 10.1016/j.lungcan.2015.10.004 [DOI] [PubMed] [Google Scholar]
- 22.Malapelle U, de Rosa N, Rocco D, et al. EGFR and KRAS mutations detection on lung cancer liquid-based cytology: a pilot study. J Clin Pathol 2012;65:87-91. 10.1136/jclinpath-2011-200296 [DOI] [PubMed] [Google Scholar]