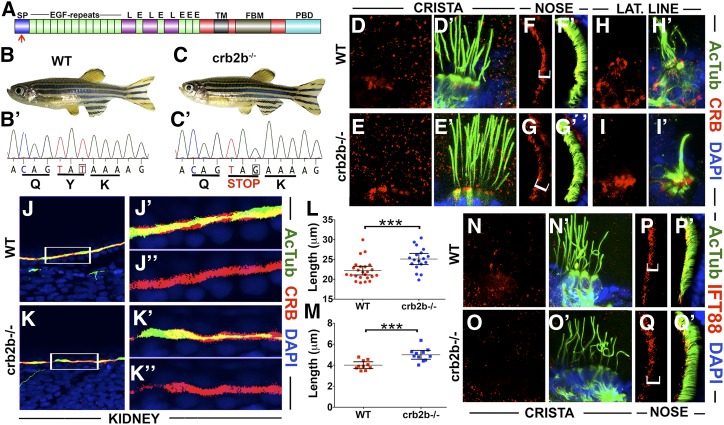
Figure 7.
crb2b affects cilia length. (A) Schematic of Crb2b protein domain structure (not to scale). Indicated are the signal peptide (SP), EGF-like repeats (E), Laminin G domains (L), transmembrane domain (TM), FERM-binding motif (FBM), and PDZ-binding domain (PBD). Red arrow shows the position of mutation in the crb2b−/−sa18042 mutant allele. (B and C) Phenotypes of wild-type (WT) (B) and crb2b−/−sa18042 (C) homozygous mutant adult zebrafish. (B’ and C’) Sequences of WT and crb2b−/−sa18042 mutant alleles. (D–I’) Whole-mount staining of WT and crb2b−/− mutants using anti-acetylated tubulin (AcTub) (green), and anti-CRB (red) antibodies at 5 days postfertilization. Samples were counterstained with DAPI to visualize nuclei (in blue). Crumbs proteins are not detected in the cilia of cristae (D–E’) and the lateral (LAT.) line (H–I’) of crb2b−/− mutants or their WT siblings. No differences in Crumbs signal are found between WT and mutants in the cilia of olfactory placodes (F–G’) and the pronephric duct (J–K’’). (L) Graph of cilia length in WT and crb2b−/− mutants. Each dot represents the average length of all cilia in one crista. Data were collected from three independent experiments using at least five animals per experiment. (M) Graph of cilia length in olfactory placodes of WT and crb2b−/− mutants. (N–O’) IFT proteins are not detected in the cristae cilia of WT (N and N’) and crb2b−/− mutants (O and O’). (P–Q’) IFT88 localization is not obviously different in nasal pit cilia of WT (P and P’) and crb2b−/− mutant (Q and Q’) animals. Brackets in (F, G, P, and Q) indicate nasal cilia. In (L and M), the mean and 95% C.I. are indicated. P < 0.001 based on Student’s t-test and Mann–Whitney test.