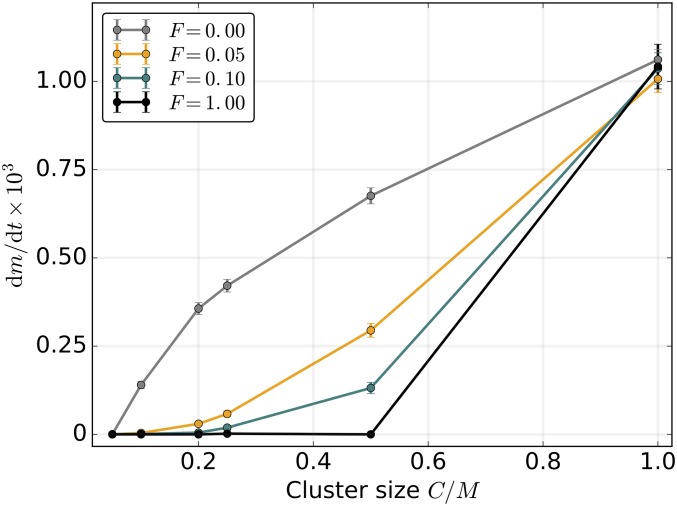
Figure 4.
Mitochondrial genome clustering promotes mutation accumulation. Nonrandom clustering of mitochondrial genomes into tightly linked groups of size C increases dm/dt due to suppression of segregational drift at cell division. However, if the clusters are allowed to exchange genomes between cell divisions, even very low migration rates F prevent the operation of Muller’s ratchet. F is the number of genome migration events per cell per generation, µ = 0.005, N = 500, M = 20, and L = 0. Here, s = 0.02 and K = 100. Error bars indicate the 95% C.I.s for the SEM ( where mfix is the number of fixed mutations after t generations).