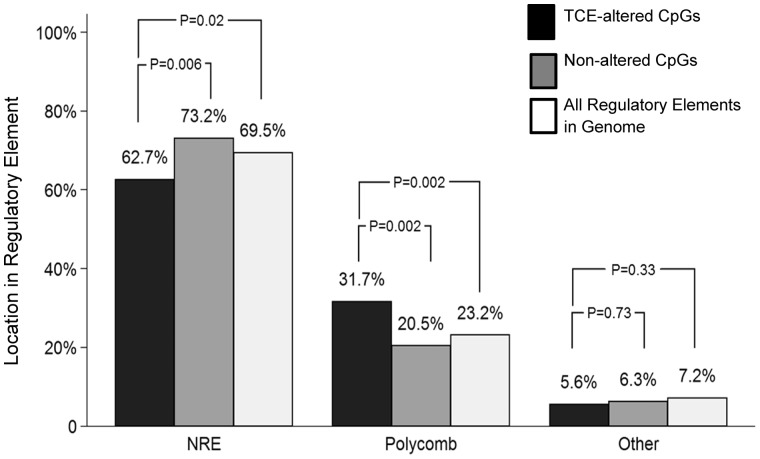
Figure 6:
Many CpG sites differentially methylated by TCE are found in PcG protein binding sites. RRBS analysis of effector/memory CD4+ T cells from control and TCE-treated mice revealed 233 DMS (methylation difference ≥ 10%, q value < 0.005). Annotation of these DMS described whether they were found outside of a transcription binding site (NRE: no regulatory element), or in a regulatory element that bound PcG proteins or other transcription factors. The percentage of CpG sites differentially methylated by TCE was compared with 400 randomly selected CpG sites not altered by TCE (Random CpGs), and to the total number of individual OREG sites known to bind Suz12, EZH2, Mtf2 or Jarid2 or other transcription factors (as identified in Mouse NCB137/mm9 genome in the UCSC Genome Browser) (All Regulatory Elements in Genome)