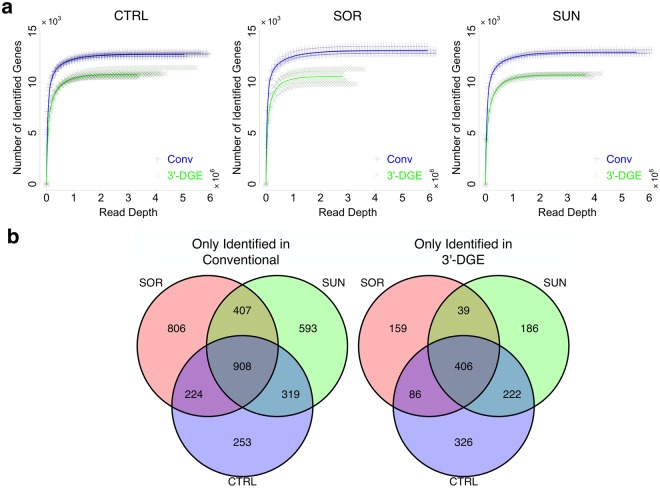
Figure 3.
Sensitivity of Gene Detection by Conventional (Conv) and 3′-end Digital Gene Expression (3′-DGE) mRNA Sequencing Methods. (a) Gene-wise reads are removed from every sample in a probability proportional to the abundance of the gene in a sample, to generate a set of the number of identified genes over a range of simulated read depths. The curves for individual replicate samples are shown with the thinner points, showing in general low variability. The average is shown with the solid line. (b) Each replicate from both mRNAseq technologies were downsampled via read removal to a common read depth (2.8 million reads per sample), and the differences in identified genes were analyzed. Most genes identified in conventional but not 3′-DGE were shared across treatment conditions, and likewise for those identified by 3′-DGE but not conventional. Specific gene names are listed in Tables S3,S4.