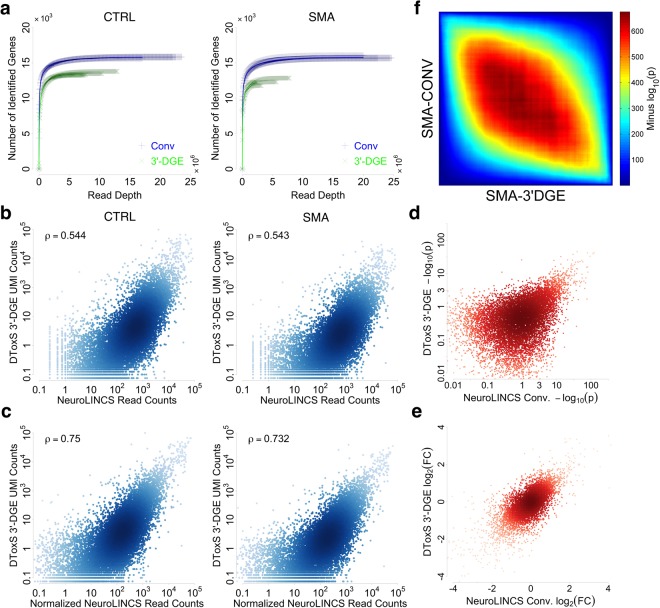
Figure 6.
Comparison between Conventional (Conv) and 3′-end Digital Gene Expression (3′-DGE) mRNA Sequencing Methods with an Independent Dataset. (a) Sensitivity of gene detection by two methods. Gene-wise reads are removed from every sample in a probability proportional to the abundance of the gene in a sample, to generate a set of the number of identified genes over a range of simulated read depths. The curves for individual replicate samples are shown with the thinner points, showing in general low variability. The average is shown with the solid line. (b,c) Quantitative gene-wise comparison between two methods. Density of points in scatter plots is indicated by depth of color. Inset text box shows Pearson correlation. In all plots, data are scaled so units are comparable. B. Scatterplots of UMI counts for DToxS’ 3′-DGE versus read counts for NeuroLINCS’ conventional, without normalization by average transcript length. CTRL or SMA refer to the genetic status of the iPS cells (see Methods). (c) Scatterplots of UMI counts for DToxS’ 3′-DGE versus transcript length-normalized read counts for NeuroLINCS’ conventional. (d,e) Comparison of statistical significance (d) or fold-change (e) for all genes identified from SMA samples by DToxS’ 3′-DGE method and NeuroLINCS’ Conv method. (d) The negative base-10 logarithm of the p-value for differential expression is plotted for each technique, with depth of color indicating density of points. (e) The log base two fold-change is plotted for each technique, with depth of color indicating density of points. (f) Rank-rank Hypergeometric tests for consistency of differential expression ranking and gene expression signatures. All genes for which a p-value for differential expression was calculated were first sorted into up or down regulated genes (as compared to CTRL), and then ranked by statistical significance. The probability of overlap between two different such rank lists was calculated with Fisher’s Exact Test (aka hypergeometric test), and visualized with a heatmap, for all combinations of list cutoffs. Shown here are lists from SMA vs. control for 3′-DGE and conventional.