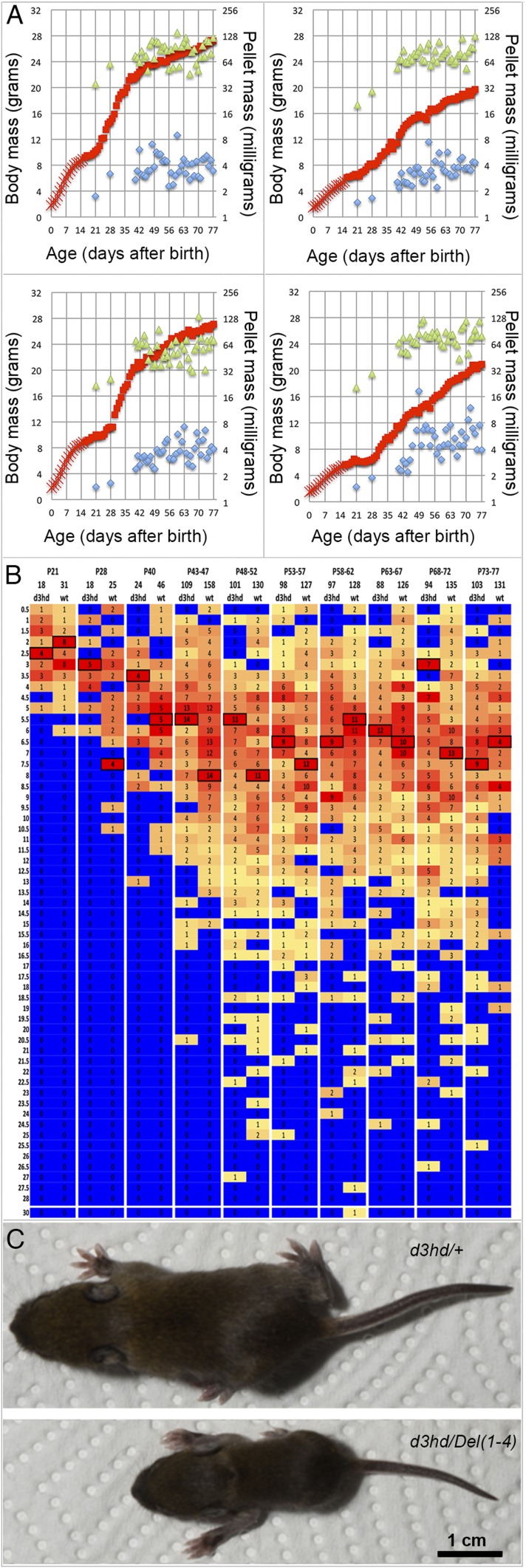
Fig. S2.
Growth retardation in F0 mice after inactivation of Hoxd3 by CRISPR/Cas9. (A) Daily body weight (red), total weight of dropping (green), and average weight (blue) are plotted for two normal siblings (Left) and two Hoxd3hd mutants (Right). All four were males issued from pronuclear injections of the Hoxd3 homeodomain-targeting plasmid vectors. (B) Frequency distributions of individual dropping masses pooled from the pairs of wild-type control and mutant individuals illustrated in A. Age at the time of collections is indicated above the heat map, total counts by wild-type and mutant genotypes are indicated as pairs below the heat map, the size bins set to 0.5-mg increments are indicated on the left. Heat maps reflect the relative frequency of pellet counts in a given lot; the most frequent values are boxed for better visibility. (C) Photograph of two P12 siblings from a litter born to a Hoxd3hd founder animal and a Del(1–4)-heterozygous male. Genotypes indicated for each specimen. A single picture was cut into two pieces for the ease of figure presentation (the scale is the same for both mice).