Significance
We present an identification of the relevance of WD40-repeat (WDR) genes in brain connectivity, highlighting the power of unbiased mouse studies in the field of neuroscience. We focus on the poorly studied WDR47 protein sharing structural homology with LIS1, which causes lissencephaly. WDR47 plays a role in progenitor proliferation, neuronal migration, and fiber tract projections in a similar fashion to LIS1 but with the distinctive particularity that WDR47 inhibits autophagic flux. This provides a functional link between autophagy biology and the C-terminal to LisH domain in mammals. Importantly, WDR47 uncovers an aspect of corpus callosum biology pointing toward a link between the regulation of microtubule dynamics and autophagic flux for axonal outgrowth and guidance.
Keywords: WD40-repeat proteins, corpus callosum agenesis, microcephaly, neurogenesis, autophagy
Abstract
The family of WD40-repeat (WDR) proteins is one of the largest in eukaryotes, but little is known about their function in brain development. Among 26 WDR genes assessed, we found 7 displaying a major impact in neuronal morphology when inactivated in mice. Remarkably, all seven genes showed corpus callosum defects, including thicker (Atg16l1, Coro1c, Dmxl2, and Herc1), thinner (Kif21b and Wdr89), or absent corpus callosum (Wdr47), revealing a common role for WDR genes in brain connectivity. We focused on the poorly studied WDR47 protein sharing structural homology with LIS1, which causes lissencephaly. In a dosage-dependent manner, mice lacking Wdr47 showed lethality, extensive fiber defects, microcephaly, thinner cortices, and sensory motor gating abnormalities. We showed that WDR47 shares functional characteristics with LIS1 and participates in key microtubule-mediated processes, including neural stem cell proliferation, radial migration, and growth cone dynamics. In absence of WDR47, the exhaustion of late cortical progenitors and the consequent decrease of neurogenesis together with the impaired survival of late-born neurons are likely yielding to the worsening of the microcephaly phenotype postnatally. Interestingly, the WDR47-specific C-terminal to LisH (CTLH) domain was associated with functions in autophagy described in mammals. Silencing WDR47 in hypothalamic GT1-7 neuronal cells and yeast models independently recapitulated these findings, showing conserved mechanisms. Finally, our data identified superior cervical ganglion-10 (SCG10) as an interacting partner of WDR47. Taken together, these results provide a starting point for studying the implications of WDR proteins in neuronal regulation of microtubules and autophagy.
The function of WD40-repeat (WDR)-containing proteins, one of the largest eukaryotic protein families, is largely unknown. Their importance is, however, evident based on their highly conserved repeating units from bacteria to mammals (1), commonly made of seven repetitive blades of 40 amino acids that end with a tryptophan-aspartic acid dipeptide at the C terminus.
As shown by crystallography studies, including the crystal structure of the beta gamma dimer of the G-protein transducin (2), a classical WDR protein, all WDR proteins are predicted to fold into a circularized beta-propeller structure, serving as a rigid platform (or scaffold) for protein–protein interactions by providing many stable and symmetrical surfaces (3, 4). One reason why WDR domains may have been less studied than other common domains, such as kinases or PDZ or SH3 domains (3), is that no WDR domain has yet been found with catalytic activity (3), but this does not mean that the scaffold domains are less important. To the contrary, their serving as a platform for multiple enzymatic reactions and signaling events is highly significant (5).
In recent years, human genetic studies have also begun to recognize the importance of WDR genes. Among 286 WDR genes annotated across both human and mouse genomes, mutations in 27 WDR genes (9.4%) have so far been implicated in brain disorders, notably in intellectual disability associated with malformations pertaining to anomalies of the corpus callosum (Dataset S1). Among these, PAFAH1B1 [also known as LIS1, a WDR protein identified 20 y ago to regulate dynein activity and neuronal migration (6)] is linked with lissencephaly type 1, a severe malformation where the brain develops without convolutions (Online Mendelian Inheritance in Man 607432), and the corpus callosum is thinner (7). Mutations in WDR62 cause autosomal recessive primary microcephaly and hypoplasia of the corpus callosum (8), and WDR73 is implicated in Galloway–Mowat syndrome characterized by microcephaly and thin corpus callosum (9). Understanding the underlying pathophysiological mechanisms of callosal disorders is critical for patient stratification and therapy development.
Made of ∼190 million axonal projections, the human corpus callosum is the largest interhemispheric white matter tract in the brain, with neurons located mainly in neocortical layers II/III, giving rise to callosal axons (10). The genetics of corpus callosum biology is, however, highly heterogeneous, and despite technological advances in next generation sequencing, 75% of callosal disorders have no identified genetic cause (11). Recent studies have suggested that a smaller corpus callosum is associated with a higher risk for autisms (12), bipolar disorder (13), and schizophrenia (14). Corpus callosum abnormalities are often seen in conjunction with other defects, such as smaller or larger brain size and malformations of cortical development (15). The formation of the corpus callosum is a process relying on axonal guidance cues, such as Netrin/DCC, ROBO, and Slit (16). This developmental process also relies on microtubule polymers that localize to the tip of the axon, known as the growth cone (17). However, much less is known about microtubules at the growth cone, but they are the primary effectors of axonal movement and guidance (18).
Less than 3% of WDR proteins have been functionally defined in the CNS, while for the remaining 97%, the function remains completely unknown (Dataset S1). Interestingly, several WDR proteins have been linked to microtubules in KO mouse studies [for example, LIS1 (19) and WDR62 (20)]. Microtubules are critical components of the cytoskeleton, and their dynamics refers to the continuous remodeling between assembly (rescue) and disassembly (catastrophe) at their tip (21). Proper regulation of this dynamic is essential and achieved through microtubule-associated proteins.
In this study, we ask whether microtubule-mediated processes might be affected by mutations of WDR genes that result in corpus callosum anomalies, what the underlying cellular and molecular mechanisms are, and ultimately, how these underlie corpus callosum biology.
Results
Mouse WDR Proteins Are Implicated in Corpus Callosum Biology.
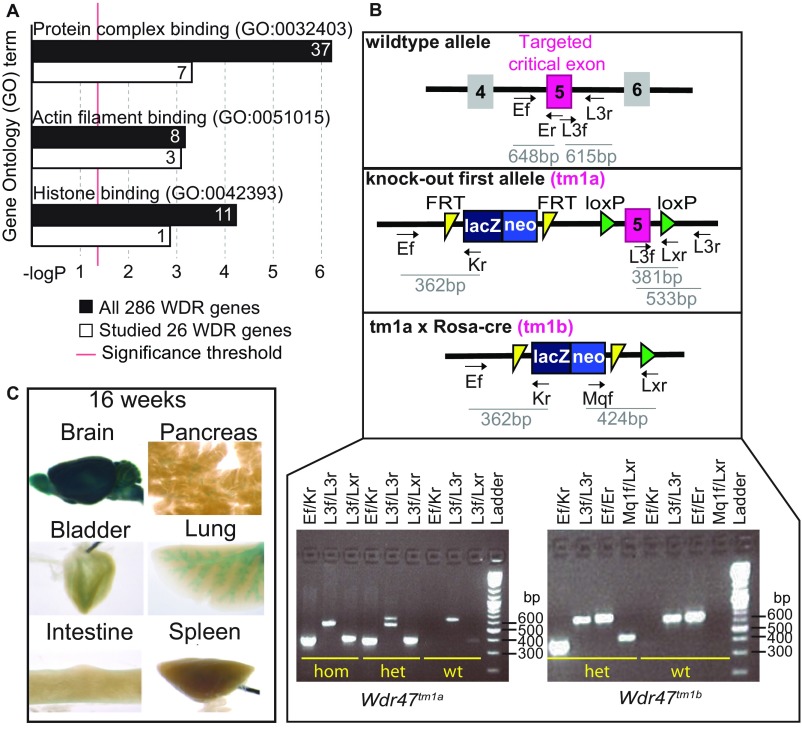
Twenty-six WDR domain-containing mouse mutants were randomly selected among a manually curated list of 286 family members (Dataset S1) and studied at 16 wk of age. We first carried out gene ontology enrichment analysis in both 286- and 26-gene sets and found the same three most significant terms (P < 0.001): protein complex binding, actin filament binding, and histone binding (Fig. S1A).
Fig. S1.
Gene ontology (GO) term analysis, Wdr47 genotyping strategy, and LacZ profiling. (A) Gene enrichment analysis of molecular function for 286 hand-curated WDR murine genes vs. 26 WDR genes analyzed in this study. x Axis shows the negative logarithm (with base 10) of the P value, and y axis represents the GO term. (B) Allelic construction of KO mouse models and genotyping strategy; tm1a refers to the KO-first allele, and tm1a crossed with ROSA Cre deleter produces tm1b (complete KO). The illustration locates the seven primers designed to specifically target each component of the construction. Expected amplicon size is shown below each primer pair. A PCR example of the primer combinations used to genotypes Wdr47tm1a and Wdr47tm1b mice is shown in Lower Right. (C) Adult mice LacZ expression patterns across a selection of six tissues (brain, pancreas, bladder, lung, intestine, and spleen) accessible through the International Mouse Phenotyping Consortium (www.mousephenotype.org/data/genes/MGI:2139593) website.
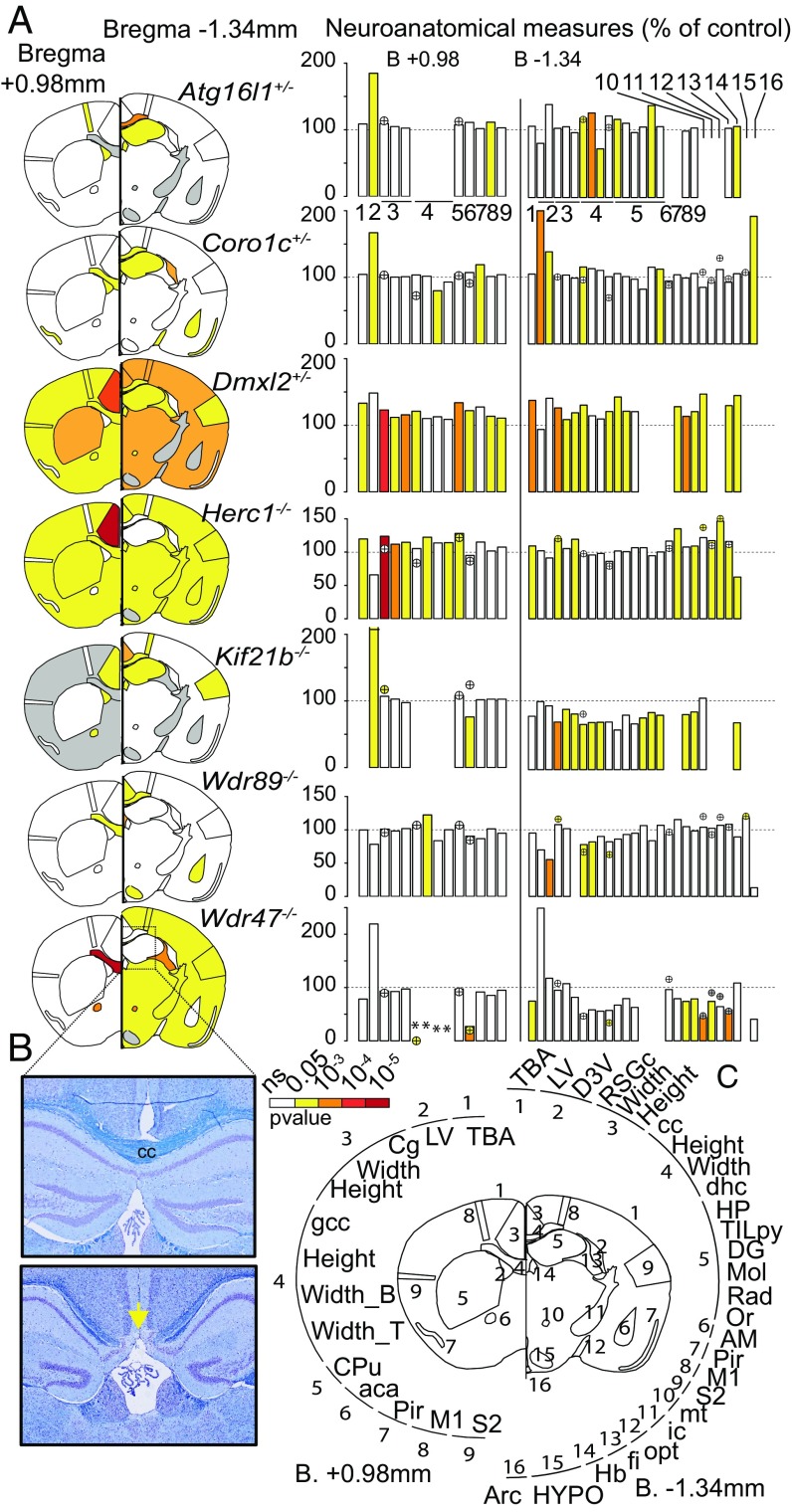
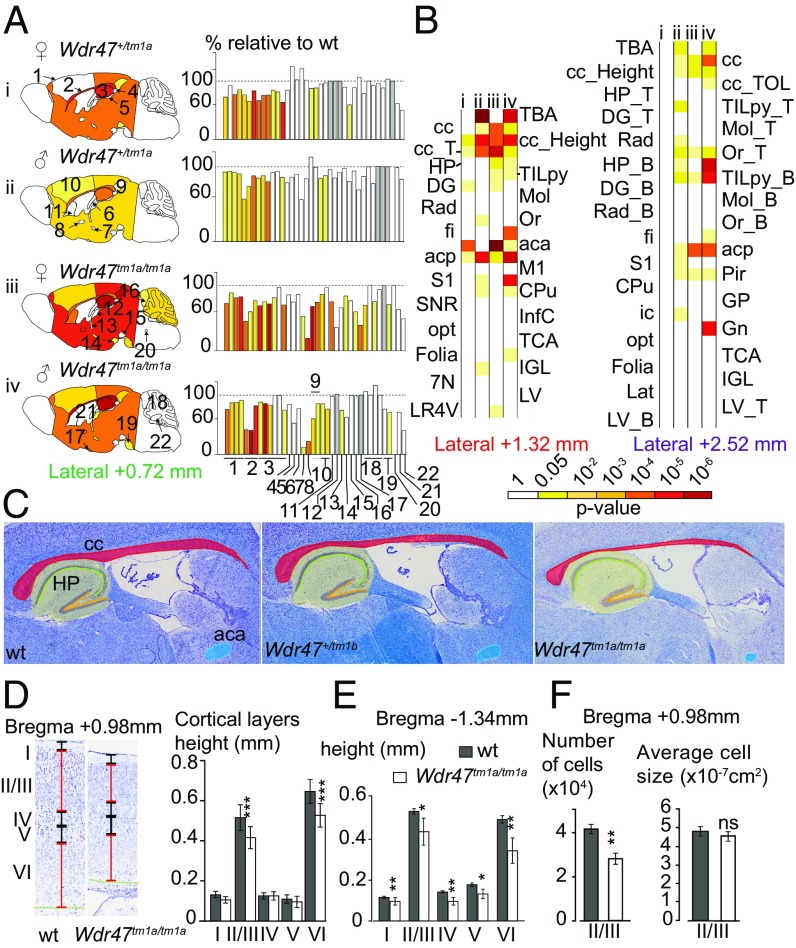
Using a quantification approach of 66 morphological and 115 cellular measurements across 19 different brain regions in two histological sections at Bregma +0.98 mm and –1.34 mm (Datasets S2 and S3), we found that mutations of seven WDR genes (Atg16l1+/−, Coro1c+/−, Dmxl2+/−, Herc1−/−, Kif21b−/−, Wdr47−/−, and Wdr89−/−) were associated with neuroanatomical phenotypes (Fig. 1; Datasets S4 and S5 show P values and percentage changes). At the morphological level, Dmxl2+/− and Herc1−/− displayed macrocephaly, with increased sizes of 37% (P = 0.0005) and 20% (P = 0.002), respectively. By contrast, Wdr47−/− and Kif21b−/− revealed microcephaly, with decreases in size of 25% (P = 0.03) and 20% (P = 0.05), respectively. Atg16l1+/− was associated with increased height (+11%) of the motor cortex (P = 0.003) and increased height (+38%) of the radiatum layer of the hippocampus (P = 0.004), whereas Wdr89−/− and Coro1c+/− were associated with ventricular atrophy (−13%) of the dorsal third ventricle (P = 0.0009) and an enlargement (+13%) of the lateral ventricles (P = 0.0002), respectively. At the cellular level, Herc1−/− displayed a 37% increase in cell numbers in the mammillothalamic tract (P = 0.003) and a 20% increase in the granular cortex (P = 0.001), while Kif21b−/− showed a decreased number of cells (−17%) in the cingulate cortex (P = 0.03).
Fig. 1.
Relevance of mouse WDR genes in adult brain morphogenesis. (A) Brain features plotted in two coronal planes according to P values for seven WDR genes (n = 3 per group). White indicates P > 0.05, and gray indicates no data. Histograms of percentage changes relative to WT animals (100%) are colored according to the significance level. Circles with crosses indicate cell count measurements. Statistical analyses were carried out using the linear mixed model framework within Phenstat (56). *Agenesis of the assessed region. (B) Brain images at Bregma −1.34 mm stained with cresyl violet and luxol blue showing the corpus callosum (cc) in WT (Upper) and agenesis of the cc in Wdr47−/− (Lower). The yellow arrow shows the agenesis (absence) of the corpus callosum. (Magnification: 20×.) (C) Numbers around the circle show assessed brain regions (a description is provided in Dataset S2). aca, anterior part of anterior commissure; AM, amygdala; Arc, arcuate nucleus; B, bottom; Cg, cingulate cortex; CPu, caudate putamen; D3V, dorsal third ventricle; DG, dentate gyrus; dhc, dorsal hippocampal commissure; fi, fimbria; gcc, genu of cc; Hb, habenula; HP, hippocampus; HYPO, hypothalamus; ic, internal capsule; LV, lateral ventricles; M1, motor cortex; Mol, molecular layer of HP; mt, mammillothalamic tract; ns, not significant; opt, optical nerve; Or, oriens layer of HP; Pir, piriform cortex; Rad, radiatum layer of HP; RSGc, retrosplenial granular cortex; S2, somatosensory cortex; T, top; TBA, total brain area; TILpy, total pyramidal cell layer.
All seven WDR mutants displayed corpus callosum anomalies (Fig. 1A and Datasets S4 and S5). Developmental mechanisms regulating the dorsoventral axes of the corpus callosum being distinctive, with pioneering axons projecting from the cingulate cortex crossing the dorsal region and neurons from the neocortex regulating formation of the ventral region (22), we quantified several regions of the corpus callosum (the genu, soma, and splenium). Dmxl2+/− and Herc1−/− strongly impacted the genu, while Atg16l1+/−, Coro1c+/−, Kif21b−/−, and Wdr89−/− affected the soma only. In addition, Atg16l1+/−, Coro1c+/−, Wdr37−/−, and Wdr89−/− exhibited cell count defects in the corpus callosum, the directionalities of which were in line with those of the morphological phenotypes. Wdr47−/− stood out as the most severely affected gene, with agenesis of the corpus callosum (Fig. 1B), defined as a failure to develop the large bundle of fibers that connect the cerebral hemispheres (11).
Wdr47 Is Highly Expressed in the Adult Brain and Is Essential for Survival in Mice.
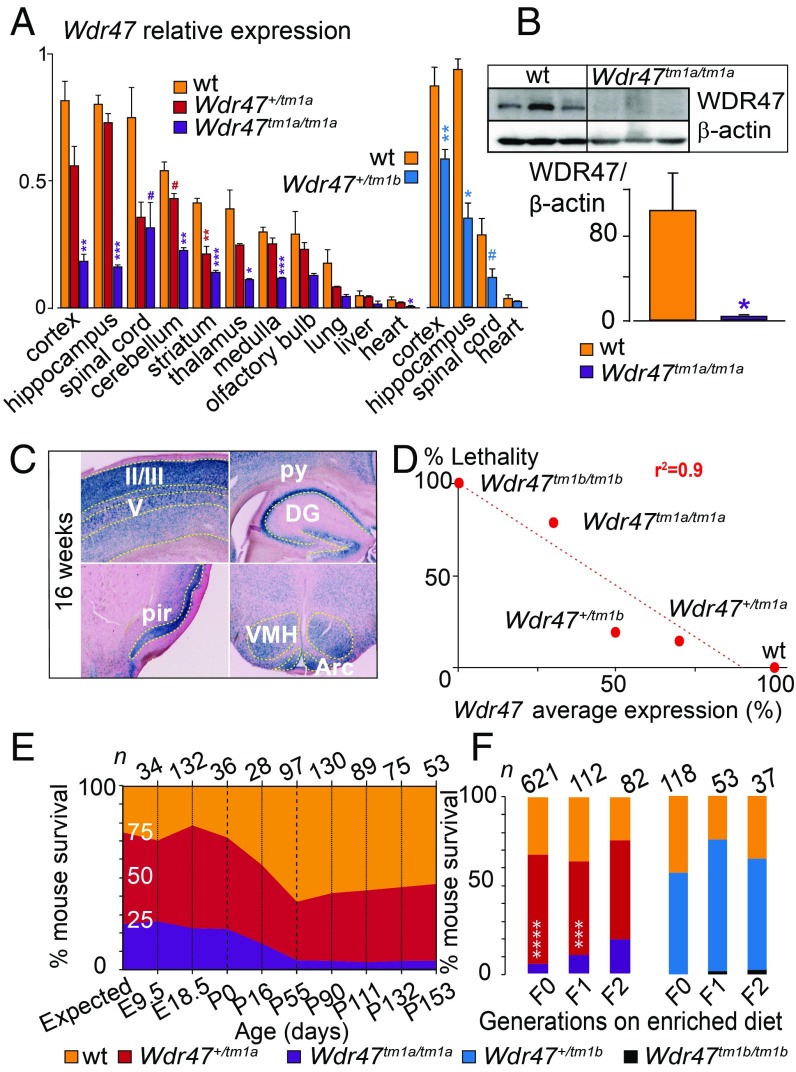
We chose to focus on the poorly studied WDR47 gene (23) given the severity of the associated neuroanatomical phenotypes and developed two mouse models (tm1a and tm1b) (Materials and Methods and Fig. S1B). We validated both models using qRT-PCR and determined that tm1a is a hypomorph allele in a series of tissues, suggesting that Wdr47 is skipping over the LacZ cassette restoring gene expression, while tm1b is a complete loss-of-function (LoF) of Wdr47 (Fig. 2A). Based on average relative expression to the WT, tm1a heterozygous (het) mice, hereafter referred as Wdr47+/tm1a, expressed 70%, and tm1b het Wdr47+/tm1b expressed 50% (Dataset S6). In homozygous (hom) animals, tm1a (Wdr47tm1a/tm1a) and tm1b (Wdr47tm1b/tm1b) expressed 30 and 0%, respectively (Dataset S6), offering the opportunity to study the impact of gene dosage (70, 50, and 30%) and complete gene LoF. WDR47 protein analysis confirmed minimal expression in Wdr47tm1a/tm1a (Fig. 2B). LacZ spatial expression throughout the brain and in peripheral tissues revealed Wdr47 expression mainly in layers II/III of the cortex, pyramidal cells of the hippocampus, spinal cord, ventromedial hypothalamus, and arcuate nucleus (Fig. 2C). Wdr47 was less expressed in peripheral tissues (Fig. 2A and Fig. S1C).
Fig. 2.
Characterization of Wdr47 mouse models. (A) Wdr47 relative expression using qRT-PCR in Wdr47+/tm1a (n = 3), Wdr47tm1a/tm1a (n = 3), and WT (n = 3) across 11 tissues and in Wdr47+/tm1b (n = 3) and WT (n = 3) across 4 tissues (cortex, hippocampus, spinal cord, and heart). Normalization was done using GNAS (guanine nucleotide-binding protein, alpha-stimulating). (B) WDR47 protein profiling in cortex of WT (n = 3) and Wdr47tm1a/tm1a (n = 3). Normalization was done using β-actin. (C) LacZ staining in adult Wdr47+/tm1a across the cortex, pyramidal cells (py), dendate gyrus (DG), piriform cortex (pir), arcuate nucleus (Arc), and ventromedial part (VMH) of the hypothalamus. (Magnification: 20×.) (D) Correlation between Wdr47 average expression and percentage mouse lethality; 843 Wdr47tm1a and 242 Wdr47tm1b were used. A linear regression was fitted (r2 = 0.9). (E) Mouse survival outcome carried out at nine time points both in Wdr47tm1a males and in Wdr47tm1a females. Expected ratio indicates 25% for WT, 50% for Wdr47+/tm1a, and 25% for Wdr47tm1a/tm1a. (F) Mouse survival outcome on supplementation in fortified diet with extra lipids and folic acid (3 vs. 0.7 mg) in Wdr47tm1a and Wdr47tm1b across three generations. Plots are represented as mean + SEM. Statistical analysis was done using Student’s t test (two-tailed; A and B) and χ2 test relative to expected counts (F). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 1E-06; #P < 0.07.
Adult mouse survival was assessed from 1,085 successfully genotyped mice derived from a heterozygous-by-heterozygous (“het-by-het”) breeding scheme (Dataset S7); 5.7% hom, 54.2% het, and 40.1% WT were obtained in Wdr47tm1a, and 0% hom, 55% het, and 45% WT were obtained in Wdr47tm1b, indicating lethality in both het and hom mice. Wdr47 expression levels and lethality (expressed as percentages) exhibited a high negative correlation (Fig. 2D), with males and females being equally affected. To determine the window of death, we then tested mouse viability from embryonic d 9.5 (E9.5) to 153 d of age (P153) (Fig. 2E). Death rate was unaffected during embryogenesis, and no abnormality in number of somites, limb morphology, and heartbeat was observed in E9.5 embryos (n = 34) and E18.5 Wdr47tm1a/tm1a embryos (n = 132). However, the percentage of Wdr47tm1a/tm1a decreased exponentially from birth to P55, with a reduction of 36% by P16 and a further reduction of 64% by P55. Mice that survived until P55 survived until adulthood. The cause of lethality remains unknown; however, histological assessment at E18.5 excluded lung defects.
It has been recently reported that a lipid-enriched diet rescues lethality in a mouse model of amyotrophic lateral sclerosis (24). We thus maintained a separate colony of mice on a fortified diet (Mouse Breeder Diet 5021) with extra lipids (10.8% as opposed to 3% in a normal diet) and folic acid (3 vs. 0.7 mg) using a het-by-het breeding scheme and examined its effects on 591 mice in both Wdr47tm1a and Wdr47tm1b (Fig. 2F). Remarkably, we found an almost complete transgenerational rescue of the lethal phenotype at the second generation in Wdr47tm1a mice. These results indicate that diet enrichment counterbalances the lethality effect, possibly by altering nutrient levels necessary in key processes for survival. There was no rescue in Wdr47tm1b, suggesting that residual Wdr47 expression is necessary for diet-induced survival reversal.
Wdr47 Deficiency Results in Severe Microcephaly and Fiber Tract Hypoplasia in Adult Male and Female Mice.
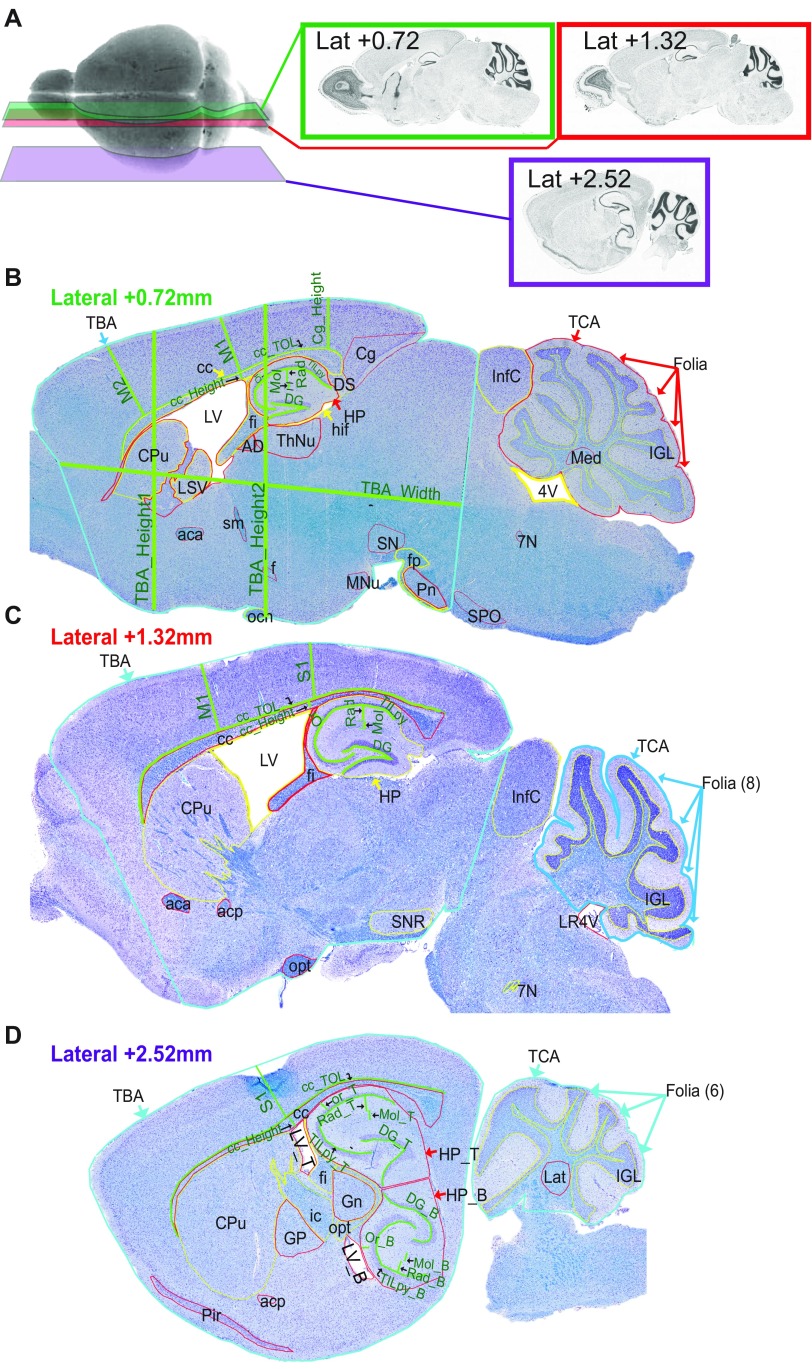
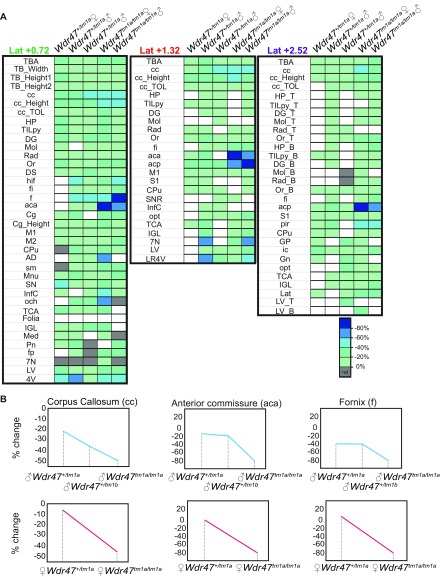
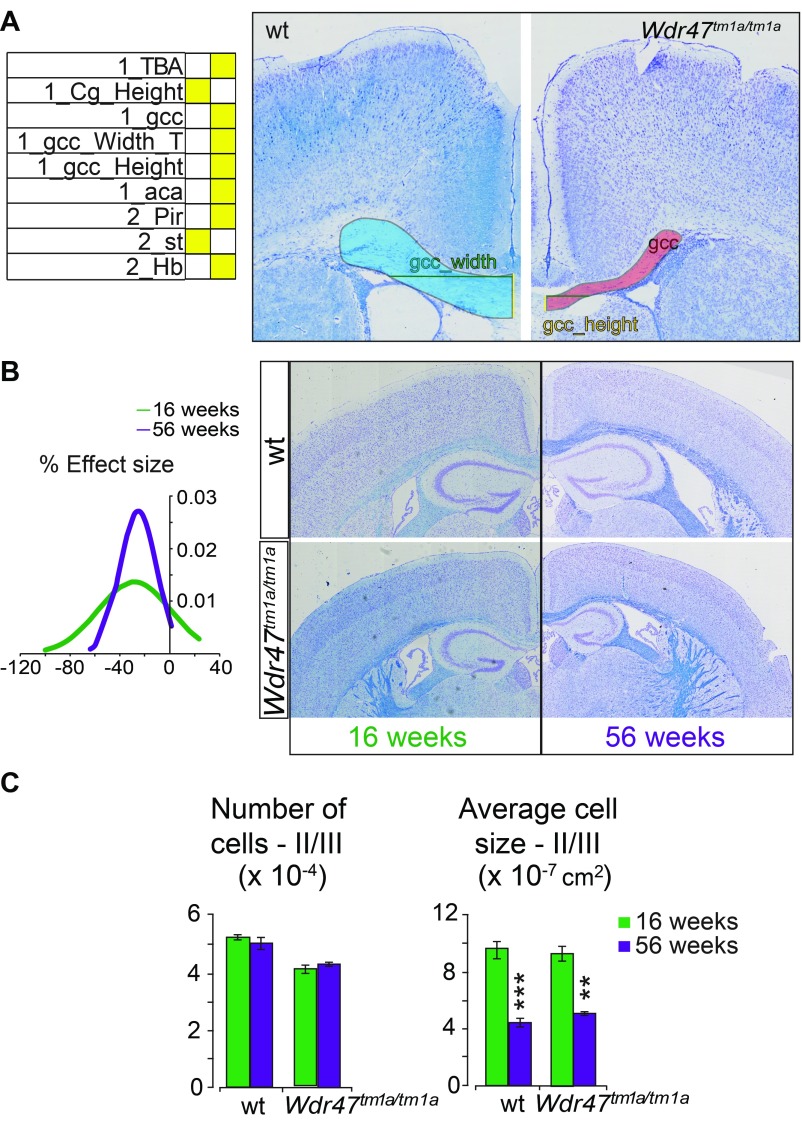
Sexual dimorphism was assessed through a newly designed sagittal analysis of 95 variables for 22 unique brain regions across three selected sections (lateral 0.72, 1.32, and 2.52 mm) (Fig. S2). This analysis had the advantage of adding new brain regions (such as the substantia nigra) while maintaining existing ones in 16-wk-old Wdr47+/tm1a and Wdr47tm1a/tm1a (Datasets S8 and S9). Consistently, male and female Wdr47tm1a/tm1a showed a similar set of neuroanatomical anomalies (for example, at lateral 0.72 mm), a reduction in the total brain area of 27.5% for female (P = 0.014) and 25.1% (P = 0.0015) for male, a decrease in the area of the corpus callosum of 54.9% (P = 0.027) for female and 54.8% (P = 0.046) for male, and a smaller anterior commissure area of 80.9% (P = 0.0007) for female and 75.2% (P = 0.004) for male (Fig. 3A). Conservatively, these results were recapitulated at lateral 1.32 and 2.52 mm, and Wdr47+/tm1a showed similarities in neuroanatomical phenotypes but to a lesser extent than Wdr47tm1a/tm1a (Fig. 3 A and B and Fig. S3A). Interestingly, we found a strong correlation between Wdr47 relative expression and severity of brain structural anomalies, such as in the corpus callosum and anterior commissure (Fig. 3C), both in male and female, showing that the role of Wdr47 in brain morphogenesis is highly sensitive to dosage (Fig. S3B).
Fig. S2.
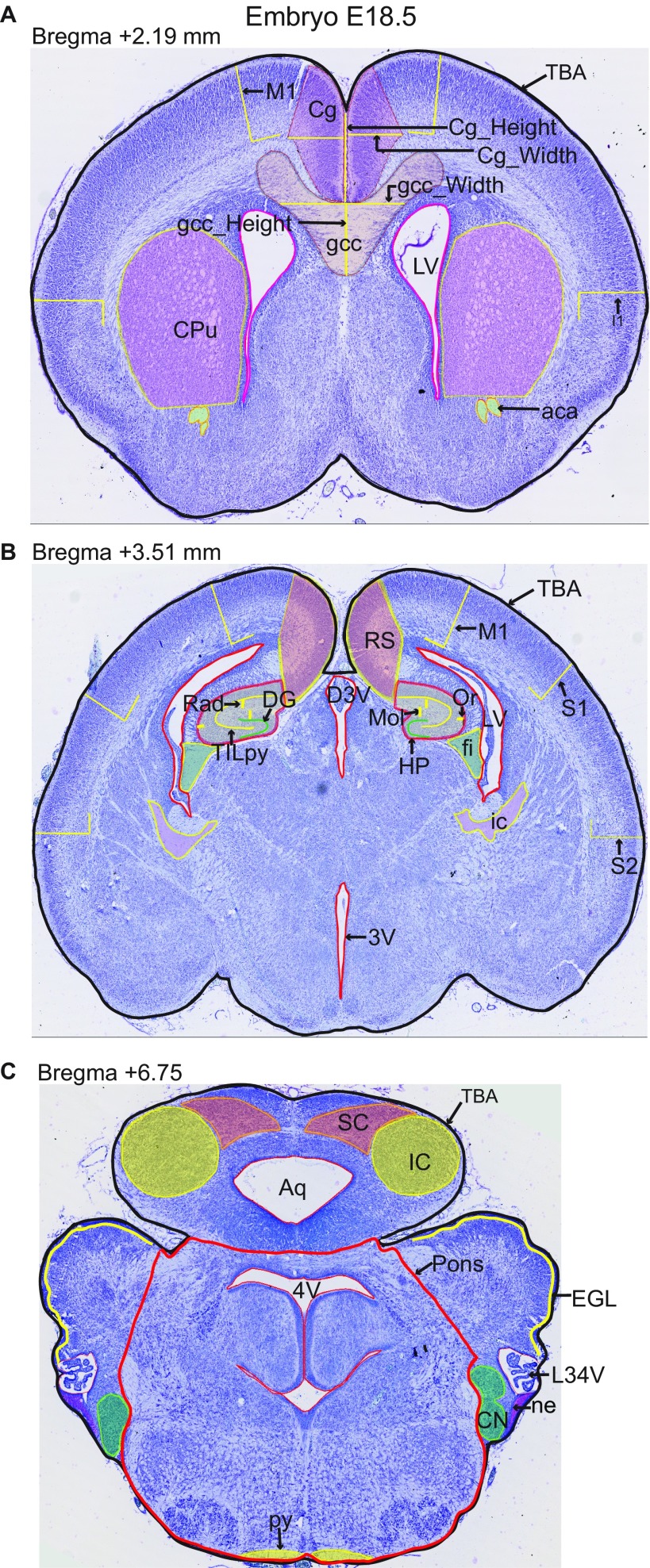
Sagittal sections of interest in adult mice (16 wk of age). (A) Representative image of histological workflow and specific lateral positions at which sagittal sections were cut for quantitative analysis. (B) Regions quantified using ImageJ have been traced on the representative image of the sagittal section 7 of interest lateral 0.72 mm. (C) Representative image of sagittal section 8 of interest (lateral 1.32 mm). (D) Representative image of sagittal section 9 of interest (lateral 2.52 mm). All brain sections were stained using cresyl violet and luxol blue. 4V, fourth ventricle; 7N, facial nucleus; aca, anterior part of anterior commissure; acp, posterior part of anterior commissure; AD, anterodorsal thalamic nucleus; B, bottom; cc, corpus callosum; Cg, cingulate cortex; Cpu, caudate putamen; DG, dentate gyrus; DS, dorsal subiculum; f, fornix; fi, fimbria; Folia, number of folia; fp, fibre of pons; Gn, geniculate nucleus; GP, globus pallidus; hif, hippocampal fissure; HP, hippocampus; ic, internal capsule; IGL, internal granular layer of cerebellum; InfC, inferior colliculus; Lat, lateral cerebellar nucleus; LR4V, fourth ventricle; LSV, ventral part of lateral septal nucleus; LV, lateral ventricle; M1, primary motor cortex; M2, secondary motor cortex; Med, medial cerebellar nucleus; Mn, mammilary nucleus; Mol, molecular layer of HP; och, optic chiasm; opt, optic tract; Or, oriens layer of HP; Pir, piriform cortex; Pn, pontine nuclei; Rad, radiatum layer of HP; S1, primary somatosensory cortex; sm, stria medullaris; SN substantia nigra; SNR, substantia nigra region; T, top; TB_Height1, height at Bregma +0.86 mm; TB_Height2, height at Bregma −1.34 mm; TB_Width, width of the total brain; TBA, total brain area; TCA, total cerebellar area; TILpy, total internal length of pyramidal cell layer of HP; TOL, total outer length. (Magnification: 20×.)
Fig. 3.
Major fiber tracts defects and microcephaly in adult male and female mice. (A) Heat map of 22 brain regions quantified at lateral 0.72 mm (Fig. S2B and Dataset S8) across Wdr47+/tm1a and Wdr47tm1a/tm1a, both male and female, vs. respective WT (n = 3 in each group). Histograms of percentage changes in comparison with WT (100%). (B) Heat map of 25 and 31 sagittal brain regions quantified at lateral 1.32 mm and 2.52 mm in male and female Wdr47+/tm1a and Wdr47tm1a/tm1a, respectively. (C) Sagittal sections stained with cresyl violet and luxol blue in mice with reducing relative expression of Wdr47. (Magnification: 20×.) (D) Height of cortical layers in adult Wdr47tm1a/tm1a (n = 6) compared with WT (n = 6) at Bregma +0.98 mm. (E) Height of cortical layers at Bregma −1.34 mm in Wdr47tm1a/tm1a compared with WT. (F) Number of cells and cell sizes in layers II/III in Wdr47tm1a/tm1a (n = 6) compared with WT (n = 6). Plots are represented as mean + SEM. *P < 0.05 (Student’s t test, two-tailed); **P < 0.01 (Student’s t test, two-tailed); ***P < 0.001 (Student’s t test, two-tailed). 7N, facial nucleus; aca, anterior part of anterior commissure; acp, posterior part of anterior commissure; B, bottom; cc, corpus callosum; CPu, caudate putamen; DG, dentate gyrus; fi, fimbria; Gn, geniculate nucleus; GP, globus pallidus; HP, hippocampus; ic, internal capsule; IGL, internal granule cell layer; InfC, inferior colliculus; Lat, lateral cerebellar nucleus; LR4V, fourth ventricle; LV, lateral ventricles; M1, motor cortex; Mol, molecular layer of HP; ns, not significant; opt, optical nerve; Or, oriens layer of HP; Pir, piriform cortex; Rad, radiatum layer of HP; S2, somatosensory cortex; SNR, substantia nigra; T, top; TBA, total brain area; TCA, total cerebellar area; TILpy, total pyramidal cell layer.
Fig. S3.
Comparison between male and female Wdr47 mice using sagittal sections. (A) Correlation between phenotypic severity in fornix (f), anterior commissure anterior part (aca), and corpus callosum (cc) and percentage relative expression in both male and female. (B) Heat map of the effect size of the 95 neuroanatomical phenotypes (Dataset S8) quantified on three selected sagittal sections in three KO models (n = 3, male, female); nd refers to no data. 4V, fourth ventricle; 7N, facial nucleus; aca, anterior part of anterior commissure; acp, posterior part of anterior commissure; AD, anterodorsal thalamic nucleus; B, bottom; cc, corpus callosum; Cg, cingulate cortex; Cpu, caudate putamen; DG, dentate gyrus; DS, dorsal subiculum; f, fornix; fi, fimbria; Folia, number of folia; fp, fibre of pons; Gn, geniculate nucleus; GP, globus pallidus; hif, hippocampal fissure; HP, hippocampus; ic, internal capsule; IGL, internal granular layer of cerebellum; InfC, inferior colliculus; Lat, lateral cerebellar nucleus; LR4V, fourth ventricle; LSV, ventral part of lateral septal nucleus; LV, lateral ventricle; M1, primary motor cortex; M2, secondary motor cortex; Med, medial cerebellar nucleus; Mn, mammilary nucleus; Mol, molecular layer of HP; och, optic chiasm; opt, optic tract; Or, oriens layer of HP; Pir, piriform cortex; Pn, pontine nuclei; Rad, radiatum layer of HP; S1, primary somatosensory cortex; sm, stria medullaris; SN substantia nigra; SNR, substantia nigra region; T, top; TB_Height1, height at Bregma +0.86 mm; TB_Height2, height at Bregma −1.34 mm; TB_Width, width of the total brain; TBA, total brain area; TCA, total cerebellar area; TILpy, total internal length of pyramidal cell layer of HP; TOL, total outer length.
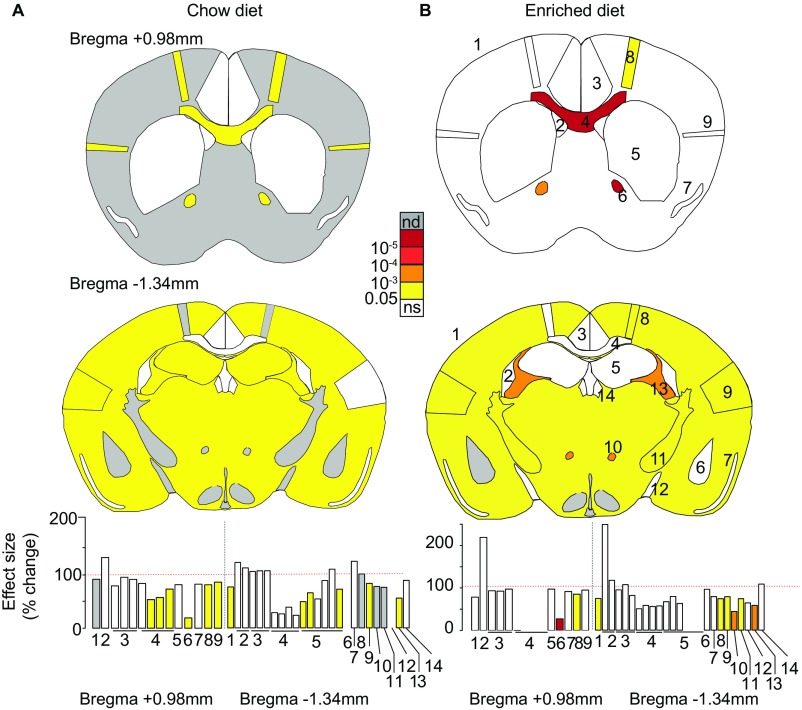
Next, we asked whether adult Wdr47 KO mice bred on an enriched diet would show reversal of neuroanatomical defects, since folic acid diet supplementation can promote neuronal proliferation and reduce apoptosis (25). For this purpose, we used the same set of 181 parameters as our original analysis of coronal sections (Dataset S2), assessed 16-wk-old Wdr47tm1a/tm1a mice derived from the second generation bred with fortified diet as well as from the chow diet colony, and compared them with their respective WTs. Similar neuroanatomical defects were identified in fortified mice compared with chow diet mice (Fig. S4), suggesting no amelioration aside from viability tests. Accordingly, additional tests on cortical layers revealed similar decrease in the height of layers II/III (−19.4%, P = 0.00007) and VI (−18.5%, P = 0.00036) at Bregma +0.98 mm (Fig. 3D) and reduction of all layers at Bregma −1.34 mm (Fig. 3E) in both diets. Focusing on layers II/III, where neurons giving rise to callosal axons originate (10), we found a 22.2% reduction in cell number (P = 0.00057) in both groups (Fig. 3F).
Fig. S4.
Impact of enriched diet on brain anatomy. Schematic representation of brain features plotted in two coronal planes according to P values for chow diet- (A) and enriched diet-fed (B) mice. The first schematic image represents the striatum section (Bregma +0.98 mm), and the second represents the hippocampus section (Bregma −1.34 mm). White indicates P value higher than 0.05, and gray is no data (nd) for absence of data. Histograms of percentage changes relative to WT animals (100%) are colored according to the significance level are shown at the bottom. Section 1–1: 1_TBA; 2: 1_LV; 3: 1_Cg, 1_Cg_Width, 1_Cg_Height; 4: 1_gcc, 1_gcc_Height, 1_gcc_Width_T, 1_gcc_Width_B; 5: 1_CPu; 6: 1_aca; 7: 1_Pir; 8: 1_M1; 9: 1_S2. Section 2–1: 2_TBA; 2: 2_LV, 2_D3V; 3: 2_RSGc, 2_RSGc_Width, 2_RSGc_Height; 4: 2_cc, 2_cc_Width, 2_cc_Height, 2_dhc; 5: 2_HP, 2_TILpy, 2_DG, 2_Mol, 2_Rad, 2_Or; 6: 2_AM; 7: 2_Pir; 8: 2_M1; 9: 2_S2; 10: 2_mt; 11: 2_ic; 12: 2_opt; 13: 2_fi; 14: 2_Hb (a full description is in Dataset S2). ns, not significant.
Wdr47 Mice Show an Embryonic Brain Size Phenotype That Worsens at Postnatal Stages.
To discriminate primary microcephaly (defined as reduction in brain size at birth) from acquired microcephaly (when brain size is normal at birth but reduces subsequently), we studied brain morphology at E18.5 using a quantification approach of 67 measurements of size and surface (Datasets S8 and S9) across three coronal planes at stereotactic position Bregma 2.19, 3.51, and 6.75 mm (Fig. S5).
Fig. S5.
Coronal sections of interest at embryonic age E18.5. Regions quantified using ImageJ have been traced on representative images of the three coronal sections of interest. (A) Critical section 4 at 2.19 mm, (B) critical section 5 at 3.51 mm, and (C) critical section 6 at 6.75 mm. Parameter names are described in Dataset S8. All brain sections were stained using cresyl violet. 3V, 3rd ventricle; 4V, 4th ventricle; aca, anterior commissure; Aq, aqueduct; Cg, cingulate cortex; CN, cochlear nucleus; CPu, caudate putamen; D3V, dorsal 3rd ventricule; DG, dentate gyrus; EGL, external granule cell layer; fi, fimbria; Folia, number of folia; gcc, genu of corpus callosum; HP, hippocampus; I, insular cortex; IC, inferior colliculus; ic, internal capsule; LR4V, lateral recess of 4th ventricle; LV, lateral ventricules; M1, motor cortex; Mol, molecular Layer of HP; ne, neuroepithelium; Or, Oriens layer of HP; Pons, pons; py, pyramidal tract; Rad, radiatum layer of HP; RS, retrosplenial granular cortex; S1, primary somatosensory cortex; S2, secondary somatosensory cortex; SC, superior colliculus; T, top; TBA, total brain area; TILpy, total internal length of pyramidal layer. (Magnification: 20×.)
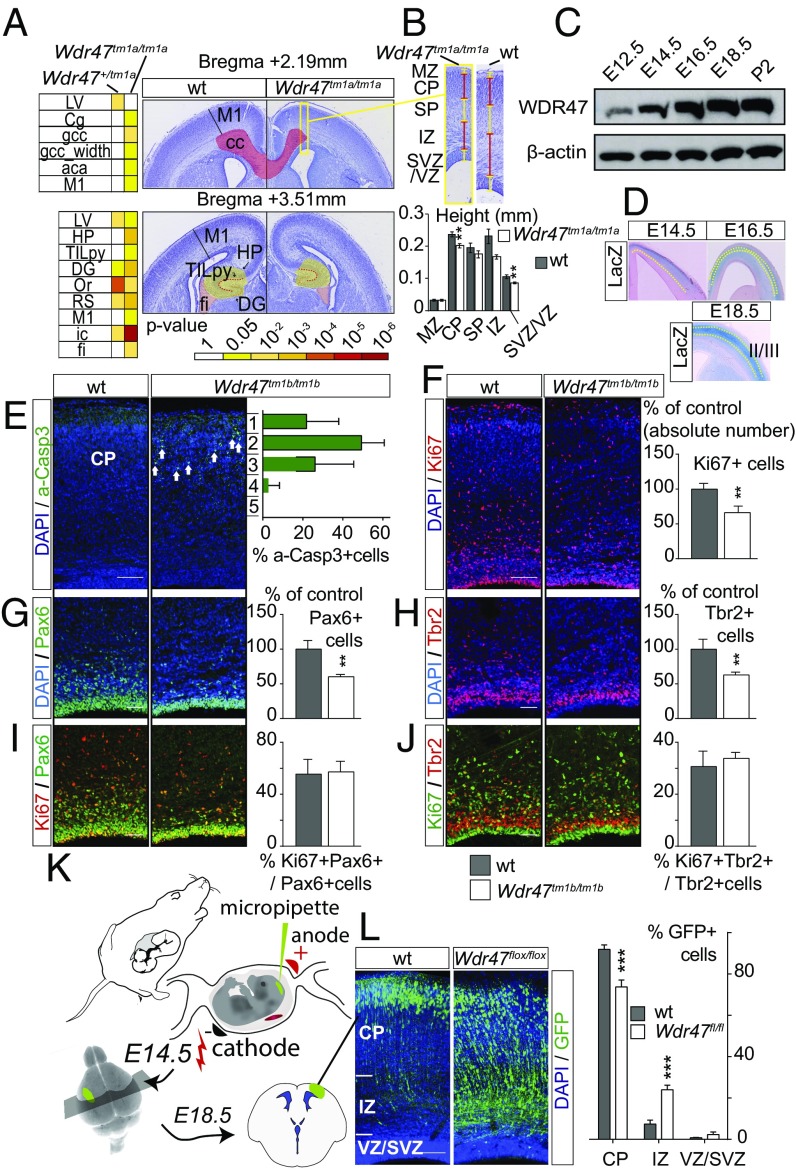
In Wdr47tm1a/tm1a, 14 phenotypes emerged as decreased, including corpus callosum area at Bregma 2.19 mm (−23.3%, P = 0.0056) and motor cortex (−14.1%, P = 0.013 and −9%, P = 0.024 at Bregma 2.19 and 3.51 mm, respectively) (Fig. 4A). While the total brain area was not significantly affected across the three coronal planes, it showed a clear tendency toward reductions of size of 10.3% (P = 0.26), 10% (P = 0.06), and 7.1% (P = 0.31) at Bregma 2.19, 3.51, and 6.75 mm, respectively (Dataset S9). Similar phenotypes emerged in Wdr47+/tm1a, with fewer regions affected and smaller percentage changes compared with Wdr47tm1a/tm1a (Fig. 4A and Dataset S9). To further investigate the reduction of the cortical thickness, we measured individual layers and found a reduction originating specifically from the cortical plate and sub- and ventricular zones at Bregma 2.19 mm (Fig. 4B).
Fig. 4.
Wdr47 is a key regulator in multiple steps of the neurogenic program. (A) Heat map of neuroanatomical defects in Wdr47tm1a at E18.5 (n = 4 Wdr47tm1a/tm1a, n = 5 Wdr47+/tm1a, n = 5 WT) (Dataset S9) and images illustrating neuroanatomical anomalies. (Magnification: 20×.) (B, Upper) Zoom in of boxed area in A showing height of neocortical layers in sections stained with cresyl violet from WT (n = 5) and Wdr47tm1a/tm1a (n = 4) embryos at E18.5. (B, Lower) Quantification of individual cortical layers. **P < 0.01 (Student’s t test, two-tailed). (C) Western blot of WDR47 expression in WT cortical tissues from E12.5 to P2. β-actin is used as a loading control. (D) LacZ expression pattern in E14.5 to E18.5 Wdr47+/tm1a across the cortex (n = 3 per group). (E) Percentage of apoptotic cells in each bin of cortical plate from E18.5 WT (n = 3) and Wdr47tm1b/tm1b (n = 3) cortices [activated caspase 3+ (a-Casp3) in green]. Arrows point to a-Casp3+ cells. (F) E18.5 WT (n = 4) and Wdr47tm1b/tm1b (n = 4) cortices showing cycling progenitors (Ki67+ in red). (G and H) E18.5 WT (n = 3) and Wdr47tm1b/tm1b (n = 3) cortices showing apical progenitors (APs; Pax6+ in green) and intermediate progenitors (IPs; Tbr2+ in red). (I and J) E18.5 WT (n = 3) and Wdr47tm1b/tm1b (n = 3) cortices showing cycling APs (Pax6+ in green and Ki67+ in red) and cycling IPs (Tbr2+ in red and Ki67+ in green). **P < 0.01 (Student’s t test, two-tailed). (K) In utero electroporation procedure. (L) E18.5 WT (n = 3) and Wdr47flox/flox (n = 3) cortices electroporated at E14.5 with NeuroD:Cre-GFP. The percentage of GFP+ cells represents neurons from the region highlighted in L. Plots are represented as mean + SEM. Images are produced using confocal microscopy, and nuclei counterstaining are performed with DAPI (blue). Wdr47tm1b/tm1b is expressed as proportion of control (F–H). aca, anterior part of anterior commissure; cc, corpus callosum; Cg, cingulate cortex; CP, cortical plate; DG, dentate gyrus; fi, fimbria; HP, hippocampus; ic, internal capsule; IZ, intermediate zone; LV, lateral ventricles; M1, motor cortex; MZ, marginal zone; Or, oriens layer; RS, retrosplenial granular cortex; SP, subplate; SVZ, subventricular zone; TILpy, total pyramidal cell layer; VZ, ventricular zone. (Scale bars: E, F, and L, 100 µm; G–J, 50 µm.) ***P < 0.0001 (two-way ANOVA followed by Bonferroni correction).
Additionally, we measured 63 parameters in Wdr47+/tm1a and Wdr47tm1a/tm1a at P8 and found a similar set of regions being affected compared with in 16-wk-old mice (Fig. S6A and Dataset S9); in particular, the total brain area was reduced by 21.6% (P = 0.046), and the corpus callosum area was smaller by 55.6% (P = 0.033). We also analyzed mice at 56 wk of age and found that the brain size phenotypes did not worsen (Fig. S6 B and C). In summary, Wdr47tm1a/tm1a mice exhibited reductions of the total brain size of 9, 22, 26, and 29% at E18.5, P8, 16 wk of age, and 56 wk of age, respectively, pointing toward primary microcephaly that worsens postnatally.
Fig. S6.
Neuroanatomical characterization of P8 and 16- and 56-wk old mice. (A) Heat map of neuroanatomical defects in Wdr47tm1a KO mice at P8 (n = 2 Wdr47tm1a/tm1a, n = 4 Wdr47+/tm1a, n = 3 WT) (Dataset S9) and representative images illustrating neuroanatomical anomalies, such as reduced primary motor cortex (M1) thickness at Bregma 2.19 mm. (Magnification: 20×.) (B) Plot of a normal distribution (based on density function) representing the effect size of 38 neuroanatomical measurements recorded on coronal plane in mice ages 56 wk old in comparison with mice at 16 wk old (male, n = 3) (Dataset S9). No visual difference is evident in the image montage of WT and Wdr47tm1a/tm1a mice coronal brain sections at 16 and 56 wk of age. (Magnification: 20×.) (C) Number and average size of cells in layers II/III of the cortex in mice ages 16 and 56 wk old (male, n = 3). All plots are represented as mean + SEM. Statistical analysis was done using Student’s t test (two-tailed). **P < 0.01; ***P < 0.001. aca, anterior part of anterior commissure; Cg, cingulate cortex; gcc, genu of the corpus callosum; Hb, habenula; Pir, piriform cortex; T, top; TBA, total brain area.
Wdr47 Regulates Progenitor Proliferation and Survival of Neurons in Late Corticogenesis.
Wdr47 cortical expression, assessed using Western blot analysis, gradually increased from E12.5 to P2, reaching a peak at E18.5 (Fig. 4C). Accordingly, LacZ spatiotemporal expression revealed an enriched Wdr47 expression in layers II/III as corticogenesis progresses (Fig. 4D), suggesting a role of WDR47 in late corticogenesis.
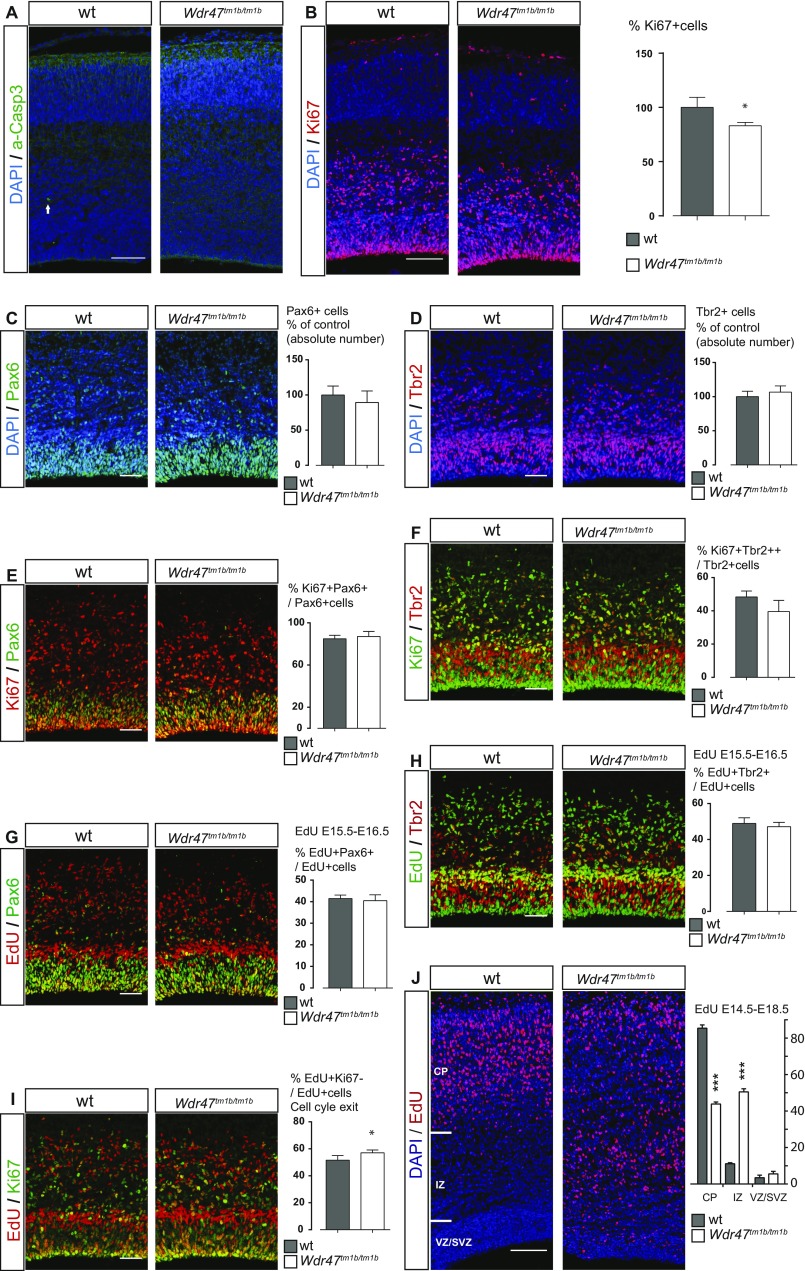
The reduction of the thickness of the cortical plate at E18.5 (Fig. 4B) could arise from a poor survival of progenitors or neurons. In agreement with this hypothesis, Wdr47tm1b/tm1b showed an increased level of apoptosis, mainly in upper-layer neurons (Fig. 4E), indicating that WDR47 is not required for the survival of the earliest born neurons. Consistently, no cell death was observed at E16.5 (Fig. S7A). We next tested whether the primary microcephaly phenotype could also stem from an impaired generation of neurons in Wdr47tm1b/tm1b embryos. Using immunolabeling, we analyzed cortical progenitors in Wdr47tm1b/tm1b and WT E18.5 embryos and found a reduced number of Ki67+ cycling progenitors (−33.8%, P = 0.0016) (Fig. 4F) and a decrease in the absolute number of both Pax6+ apical (−39.7%, P = 0.0064) (Fig. 4G) and Tbr2+ intermediate (−37.2%, P = 0.0042) (Fig. 4H) progenitors. Noteworthy, the proliferative potential of both progenitor types remained unchanged (Fig. 4 I and J).
Fig. S7.
Wdr47 deletion does not affect early stages of cortical development. (A) E16.5 WT and Wdr47tm1b/tm1b cortices showing apoptotic cells (activated Caspase3+ in green). Arrow points to a-Caspase3+ cell. (B) E16.5 WT (n = 3) and Wdr47tm1b/tm1b (n = 4) cortices showing cycling progenitors (Ki67+ in red). *P < 0.05 (Student’s t test, two-tailed). (C and D) E16.5 WT (n = 3) and Wdr47tm1b/tm1b (n = 4) cortices showing apical progenitors (C; Pax6+ in green) and intermediate progenitors (D; Tbr2+ in red). (E and F) E16.5 WT (n = 3) and Wdr47tm1b/tm1b (n = 4) cortices showing cycling apical progenitors (E; Pax6+ in green and Ki67+ in red) and cycling intermediate progenitors (F; Tbr2+ in red and Ki67+ in green). (G and H) E16.5 WT (n = 3) and Wdr47tm1b/tm1b (n = 4) cortices showing newborn apical progenitors (G; Pax6+ in green and newborn cells injected with EdU at E15.5 in red) and newborn intermediate progenitors (H; Tbr2+ in red and newborn cells injected with EdU at E15.5 in green). (I) E16.5 WT (n = 3) and Wdr47tm1b/tm1b (n = 5) cortices showing newborn cells (injected with EdU at E15.5 in red) and cycling progenitors (Ki67+ in green). *P < 0.05 (Student’s t test, two-tailed). (J) E18.5 WT (n = 3) and Wdr47tm1b/tm1b (n = 3) cortices injected with EdU at E14.5 quantified as percentage of EdU+ cells in several brain regions as indicated on the image. All plots are represented as mean + SEM. All representative images were produced using confocal microscopy, and nuclei counterstaining were performed with DAPI (blue). Wdr47tm1b/tm1b expressed as proportion of control (B–D). CP, cortical plate; IZ, intermediate zone; SVZ, subventricular zone; VZ, ventricular zone. (Scale bars: A, B, and J, 100 µm; C–I, 50 µm.) ***P < 0.0001 (two-way ANOVA followed by Bonferroni correction).
To understand the loss of progenitor cells at E18.5, we repeated our experiments at an earlier stage. Wdr47tm1b/tm1b E16.5 Pax6+ apical and Tbr2+ intermediate progenitors behaved as WT, with no observable phenotype in their absolute number (Fig. S7 C and D) or proliferative potential (Fig. S7 E and F). However, we observed a milder decrease in the number of Ki67+ cycling progenitors by 17% (P = 0.018) (Fig. S7B) compared with −33.8% at E18.5 (Fig. 4F). In addition, we assessed cell cycle exit and fate of newborn cells by injecting 5-ethynyl-2-deoxyuridine (EdU) at E15.5 and studying corresponding Wdr47tm1b/tm1b embryos 24 h later at E16.5. We found no differences in the fate of newborn Pax6+ and Tbr2+ progenitors (Fig. S7 G and H) but a slight increase in the number of progenitors that exited the cell cycle (+10.7%, P = 0.027) (Fig. S7I) together with decreased proliferation (Fig. S7B), suggesting that the reduction of progenitors self-renewal starts from E16.5 onward and progressively increases until E18.5.
Specific Deletion of Wdr47 in Postmitotic Neurons Impairs Radial Migration.
Given that WDR47 is involved in the neurogenic program and has been identified as a microtubule-associated protein (23), we tested whether, in addition to neurogenesis, WDR47 could also regulate neuronal migration. We performed acute deletion of WDR47 in projection neurons by in utero electroporation of plasmids, allowing the expression of the CRE recombinase and the GFP under the control of the NeuroD promotor (NeuroD:CRE-GFP) at E14.5 (Fig. 4K). Four days after in utero electroporation, while most of the GFP+ postmitotic neurons reached the cortical plate in the control (Fig. 4L), neurons depleted for WDR47 (NeuroD:CRE-GFP in WDR47fl/fl embryos) accumulated in the intermediate zone, with a decrease of 20% of the cells reaching the cortical plate (Bonferroni adjusted P = 0.0001) (Fig. 4L). The role of WDR47 in radial migration of projection neurons was confirmed in KO mice, as we observed similar positioning defects of EdU-labeled cells 4 d after a single EdU injection in E14.5 Wdr47tm1b/tm1b embryos compared with WT littermates (reduction of 51.3% in the cortical plate; Bonferroni adjusted P < 0.0001) (Fig. S7J). WDR47 is, therefore, required for proper radial migration of projection neurons.
Wdr47 Depletion Impairs Growth Cone Morphology and Microtubule Stability.
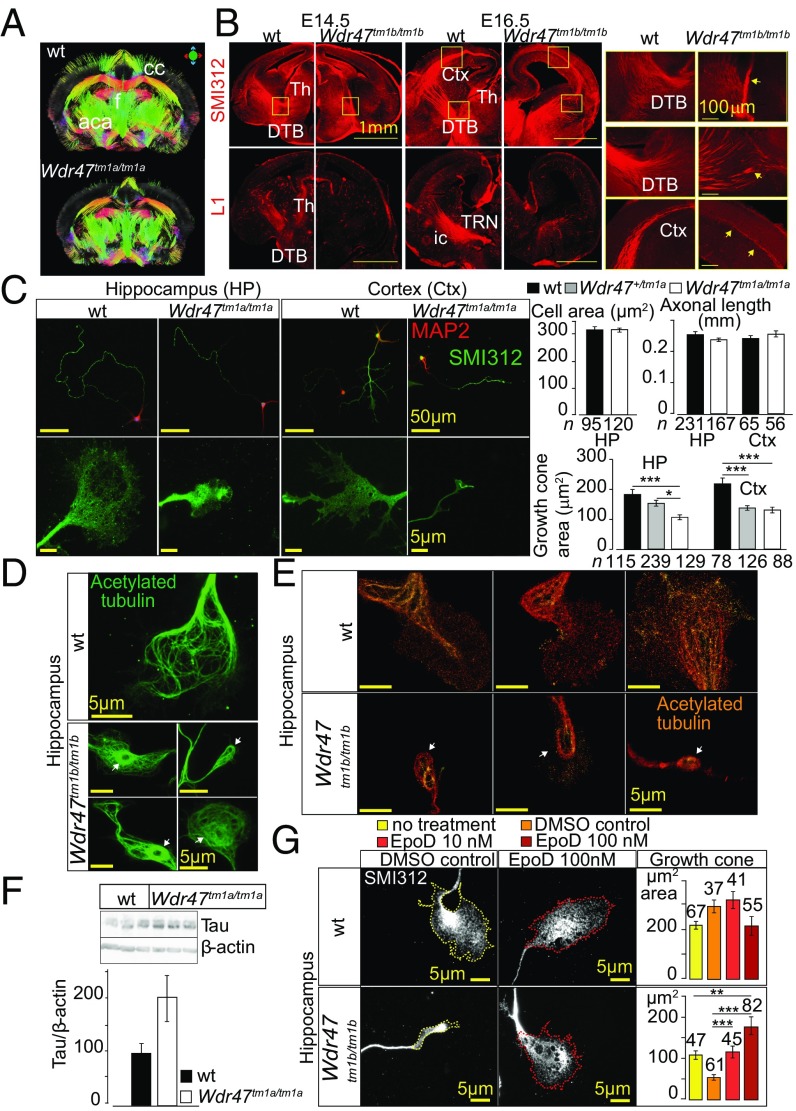
Next, we turned to the investigation of fiber projections during neurodevelopment considering the extreme hypoplasia of fiber tracts in 16-wk-old mice (Fig. 3). Consistently, MRI corroborated these findings throughout the brain, showing the corpus callosum as the most affected region (Fig. 5A). Using the axonal (SMI-312R) and L1CAM markers to visualize neurofilaments and callosal neurons, respectively, we found fewer axonal processes both at E14.5 and E16.5 Wdr47tm1b/tm1b embryos, with thalamocortical projections unable to cross the diencephalon–telencephalon boundary (Fig. 5B).
Fig. 5.
Microtubule-stabilizing role of WDR47 at the growth cone. (A) MRI in Wdr47tm1a/tm1a male. aca, anterior part of the anterior commissure; cc, corpus callosum; f, fornix. (B) Confocal microscopy images of projection patterns in the developing brain using axonal and commissural markers at E14.5 (n = 2) and E16.5 (n = 2). Ctx, cortex; DTB, diencephalon–telencephalon barrier; ic, internal capsule; Th, thalamus; TRN, thalamic reticular nucleus. (Scale bars: Left and Center, 1 mm; and Right, 100 μm.) (C) Fluorescent microscopy images of primary neurons derived from WT and Wdr47tm1a/tm1a embryos at E17.5 stained with anti-MAP2 (red) and SMI-312R (green) in hippocampal (HP) and cortical (Ctx) primary neuronal cultures. Area of cell body, length of axon, and area of growth cones were quantified using ImageJ and analyzed using the Kruskal–Wallis test. (Scale bars: Upper, 50 μm; Lower, 5 μm.) (D) Microtubule architecture studied in the growth cones of hippocampal primary neurons by staining for acetylated tubulin. White arrows show odd ring-like arrangements. (E) Superresolution single-molecule localization microscopy of hippocampal growth cone stained with acetylated tubulin in Wdr47tm1b/tm1b. (Scale bar: 5 μm.) (F) Western blot analysis of endogenous Tau levels in three Wdr47tm1a/tm1a compared with WT (n = 3). Quantification of relative protein expression is normalized against β-actin. (G) Images of primary hippocampal neurons derived from Wdr47tm1b/tm1b and WT embryos at E17.5 treated with 10 and 100 nM EpoD. Growth cone area was quantified using ImageJ after 1.5 h of treatment. (Scale bar: 5 μm.) ***P < 0.001; **P < 0.01; *P < 0.05.
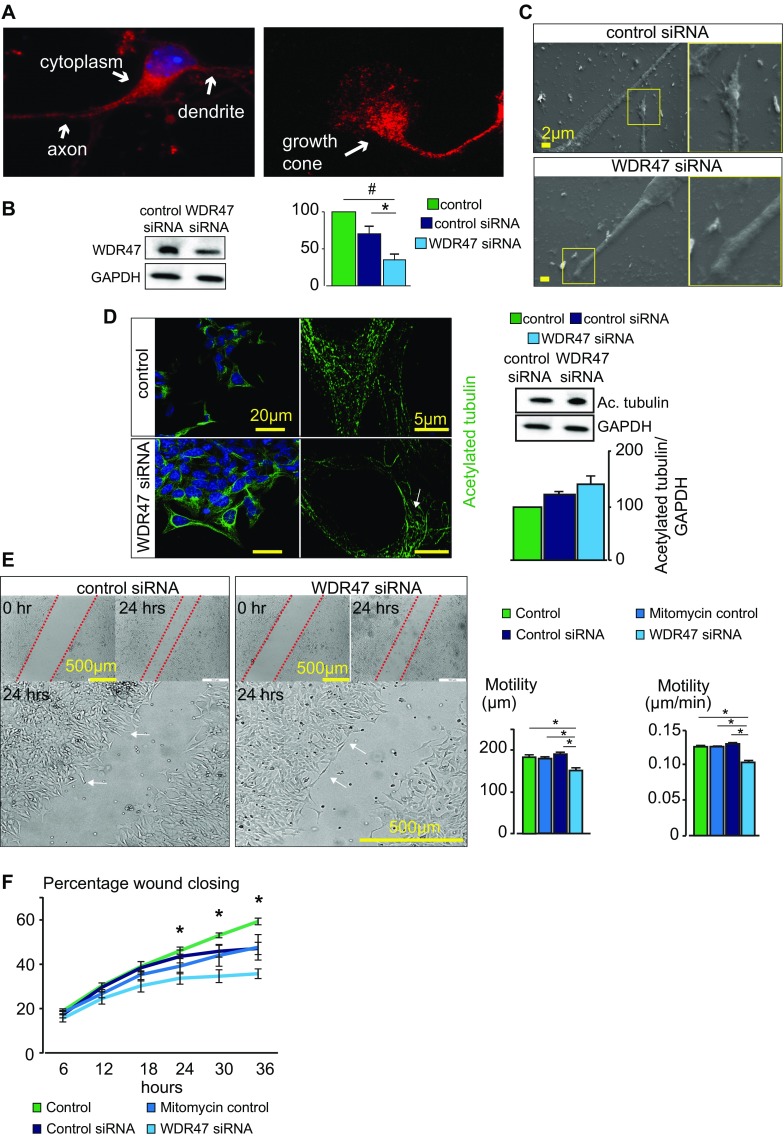
Because endogenous WDR47 is expressed throughout the cytoplasm, neurites, and in the growth cone in neurons (Fig. S8A), we thought to assess neuronal architecture when Wdr47 is depleted using cortical and hippocampal primary neuronal cultures at E17.5. We visualized the axon and growth cone using a marker for neurofilaments and microtubule-associated protein 2 (MAP2), a microtubule-associated protein enriched in the dendrites of neurons, and measured the area of the cell body (n = 215), axonal length (n = 519), and area of the growth cone (n = 775). Primary neuronal cultures derived from Wdr47tm1a/tm1a displayed a severe reduction of growth cone areas by 41% for the cortex (P = 2.69E10−5) and 42% for hippocampus (P = 1.88E10−6) (Fig. 5C), resembling the physiological collapse or catastrophe state in growth cone behavior (21). Neuronal cultures derived from Wdr47+/tm1a also showed smaller growth cone areas of 17% for the hippocampus (P = 1.08E10−5) and 37% for the cortex (P = 8.6E10−6), showing that this phenotype is also sensitive to Wdr47 dosage (Fig. 5C). Furthermore, these structures displayed a blunt tip and reduced filopodia protrusions, while the cell area and length of axon did not differ (Fig. 5C). These observations were confirmed in rat hypothalamic neurons treated with WDR47-specific siRNA using scanning EM (Fig. S8 B and C). In addition, time-lapse recordings from live neurons over 24 h showed that mutant growth cones are much less dynamic compared with WT (Movies S1 and S2).
Fig. S8.
WDR47 subcellular localization and neuronal wound assay. (A) Subneuronal localization of WDR47 using immunofluorescence (red) in primary cortical cultures of WT mice. (Magnification: 65×/one oil objective.) (B) Western blot verification of WDR47 knockdown in rat GT1-7 hypothalamus-derived neuronal cells in WDR47 siRNA-treated cells compared with control siRNA cells 24 h posttransfection (n = 7). (C) Scanning electron micrographs of control siRNA and WDR47 siRNA neurons at the level of the axon and growth cone in the migration zone. (Scale bar: 2 μm.) (D) Representative confocal (Left) and SR-SIM (Right) fluorescent micrographs of acetylated tubulin networks in control siRNA (Upper) and WDR47 siRNA-treated cells (Lower). The white arrow shows a highly convoluted structure in the perinuclear region. Western blot analysis of acetylated tubulin is carried out in control siRNA compared with WDR47 siRNA-treated cells. Normalization is done using housekeeping gene GAPDH. (Scale bars: Left, 20 μm; Right, 5 μm.) (E, Left) Transmission light micrographs of an in vitro 36-h neuronal migration assay of rat GT1-7 neuronal cells treated with control or WDR47 siRNA. The dashed red lines show the edge of the wound. White arrows indicate neurons at the edge of the wound. (E, Right) Average migration distance and migration velocity (micrometers per minute) in WDR47 siRNA-treated cells compared with the mitomycin control group or control siRNA group (n = 4 in each group). (F) Percentage of wound closure is shown over time in WDR47 siRNA-treated cells (n = 4). All plots are represented as mean ± SEM. #P < 0.07; *P < 0.05 (Student’s t test, two-tailed).
The analysis of the microtubule distribution network at the growth cone of neurons derived from Wdr47tm1b/tm1b using acetylated tubulin as a marker of stable microtubules revealed unusual shapes, with a ring-like structure at the soma (Fig. 5D). Similar abnormalities were seen in rat hypothalamic neurons using superresolution structured illumination microscopy; however, tubulin protein levels were not significantly altered (Fig. S8D). Superresolution single-molecule localization microscopy also showed these unusual shapes and further established that tubulin molecules were widely dispersed in mutant as opposed to uniform and denser distribution in WT cells (Fig. 5E). Tau protein level, a microtubule-associated protein known to modulate the stability of axonal microtubules, was increased by about twofold in Wdr47tm1a/tm1a cortical tissue samples compared with WT (Fig. 5F). Given the role of Tau in microtubule dynamics, we hypothesized that WDR47 might participate in microtubule stabilization. We tested this by treating hippocampal primary neuronal cultures derived from Wdr47tm1b/tm1b, characterized by a reduction of growth cone areas of −75% (n = 162, P = 1.89E10−5) (Fig. 5G), with a microtubule stabilizer compound [Epothilone D (EpoD)] at two concentrations (10 and 100 nM) for 1.5 h as recommended elsewhere (26). Remarkably, EpoD was able to dose-dependently rescue growth cone size up to +69.7% relative to vehicular control (DMSO) cells (Fig. 5G). Treatment with 10 nM EpoD increased the size of growth cones by 2.1 times (n = 102, P = 1.2E10−4), and treatment with 100 nM EpoD increased the size of growth cones by 3.3 times (n = 143, P = 1.9E10−9). No significant changes were observed between EpoD-treated and nontreated groups in the WT. To determine whether these anomalies might be causing cell motility defects, we used a previously tested assay in neurite outgrowth (27), which relies on creating a scratch in cell culture dishes and quantifying the time required for the cells to close it (28). At 24 h postscratch introduction, the migration distance and velocity (Fig. S8E) as well as the percentage wound closing (Fig. S8F) were reduced with WDR47 siRNA treatment. All together, these results show that Wdr47 plays a role in stabilizing microtubules, facilitating tubulin network dynamics in both genetic mutant cells and siRNA-treated cells.
Superior Cervical Ganglion-10 Is an Interacting Partner of WDR47.
To understand the molecular mechanisms by which WDR47 might regulate microtubule stability, we next searched for interacting partners by screening a human fetal cDNA library using a yeast two-hybrid system.
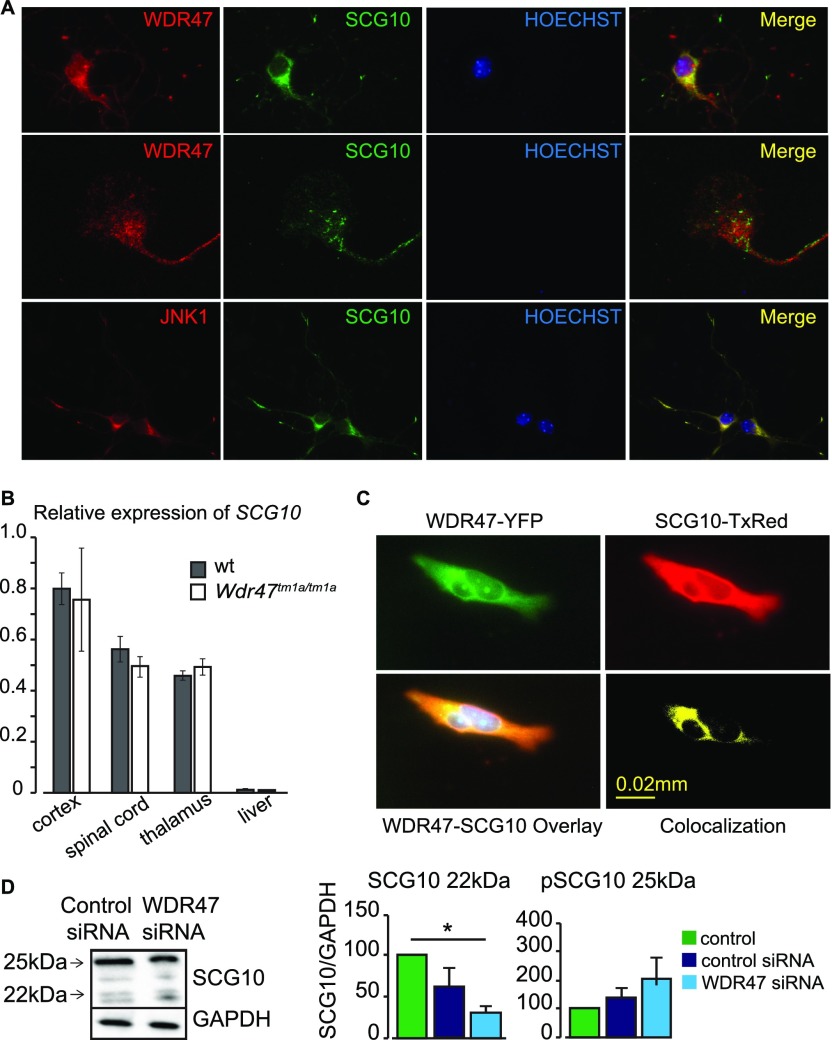
Using the N terminus of WDR47 as bait, the superior cervical ganglion-10 (SCG10) protein was identified as a putative WDR47-interacting partner (Dataset S10). SCG10 is a well-established microtubule-destabilizing protein (29) regulated by JNK1, a protein kinase of the MAPK family known to phosphorylate SCG10, rendering it inactive (30). To gain insight into the mechanistic basis of this interaction, we first studied localization of WDR47, SCG10, and JNK1 in primary cortical neurons. WDR47 colocalized with SCG10 in the cytoplasm but not in the growth cone, whereas JNK1 showed colocalization with SCG10 in the cytoplasm as well as neurites (Fig. S9A). SCG10 relative mRNA expression levels showed no difference between Wdr47tm1a/tm1a and WT mice preparations derived from the cortex, spinal cord, thalamus, and liver (Fig. S9B). Colocalization of WDR47 and SCG10 was confirmed to occur in the cytoplasm of hypothalamic cells (Fig. S9C). Western blot analysis of endogenous SCG10 normally gives rise to four bands that range from 20 to 25 kDa, representing distinct phosphorylation states (31). We assessed this in WDR47 siRNA-treated cells and quantified 22-kDa (unphosphorylated) and 25-kDa (phosphorylated) bands, but we only detected a trend for decreased 22-kDa SCG10 and increased 25-kDa SCG10 (Fig. S9D). Together, these results suggest that WDR47 physically interacts with the microtubule-destabilizing protein SCG10.
Fig. S9.
SCG10 interacts with WDR47. (A) Colocalization images of WDR47 with SCG10 and SCG10 with JNK1. (B) Relative expression of SCG10 transcripts in n = 3 Wdr47tm1a/tm1a mice using qRT-PCR compared with WT across four tissues (cortex, spinal cord, thalamus, and liver) plotted as mean + SEM. (C) Colocalization of SCG10 and WDR47 in GT1-7 hypothalamic neuronal cells. Upper Left shows expression in GT1-7 cells transfected with pEYFP-WDR47 (green). Upper Right shows expression of SCG10 labeled with Texas red anti-SCG10 antibody (red). Lower Left is the overlay of Upper, and Lower Right is the 3D colocalization of WDR47 and SCG10. (D) Western blot analysis of SCG10 relative protein levels in response to WDR47 siRNA treatment. GAPDH is used as a loading control. Statistical analysis was done using Student’s t test (two-tailed). *P < 0.05. (Magnification: A, Top and Bottom, 20×/one dry objective; A, Middle, 65×/one oil objective.)
Wdr47 Mice Are Hyperactive and Display Sensory Motor Gating Abnormalities.
To establish if the neuroanatomical defects in Wdr47 KO mice lead to specific behavioral phenotypes, we assessed a broad range of paradigms in both male and female mice (Dataset S11 shows a list of P values).
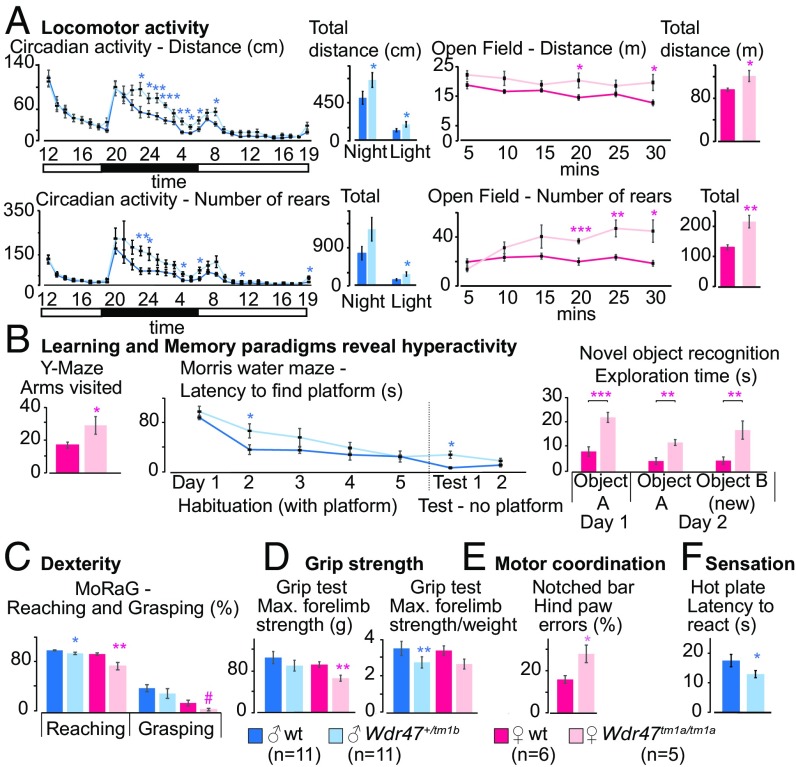
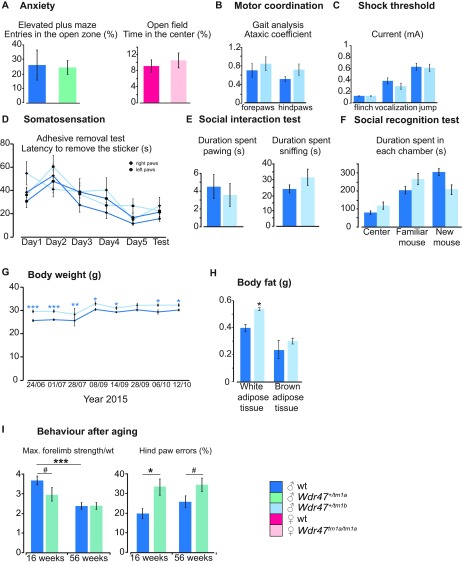
The circadian activity test revealed an increase in traveled distance (P = 0.02) during the active phase (Fig. 6A). The open-field test was used to examine basic locomotor activity as well as anxiogenic behavior and showed increased total traveled distance in the arena and total number of rears (P = 0.001) (Fig. 6A) but did not show an increase in the time spent in the center, suggesting no traits of anxiety (Fig. S10A). The elevated plus maze test confirmed the absence of anxiogenic behavior in mice (Fig. S10A). Since Wdr47 is highly expressed in the hippocampus involved in the consolidation of information, we next looked for deficits in memory using the Y-maze test to evaluate working memory, the Morris water maze for spatial memory performance, and the novel object recognition for long-term memory as well as social recognition test. Mice did not show any difficulties in learning each given task or in memorizing various objects and cues; however, they displayed, again, hyperactivity in the Y-maze (P = 0.04) and novel object recognition paradigms (P = 0.0004) (Fig. 6B). Because of prominent corpus callosum abnormalities, we studied forelimb laterality and dexterity using the Mouse Reaching and Grasping (MoRaG) test (32) and found reduced reaching (P = 0.02 for male and P = 0.005 for female) abilities, while forelimb laterality was unaffected (Fig. 6C). Accordingly, forepaws strength was decreased as studied in a grip strength test used to assess muscular strength (Fig. 6D). Hind paws motor coordination was affected in the notched bar (P = 0.01) and gait test (Fig. 6E) but not ataxia (Fig. S10B). We also tested somatosensation using the hot plate, adhesive removal (33), and shock tests. Mice showed very significant decreased latency to react to nociceptive heat (P = 0.0001), indicative of increased sensitivity (Fig. 6F). This, however, was specific to heat stimuli, since touching sensitivity and electric shock showed no difference (Fig. S10 C and D). Finally, we tested social skills using a social interaction test but found no significant differences (Fig. S10 E and F). At the metabolic level, males developed higher (+9.7%) body weight (P = 0.0001) (Fig. S10G), despite increased hyperactivity, accounted for by increased white but not brown adipose tissue (Fig. S10H). Phenotypes were replicated in a validation cohort of het male mice but were milder, suggesting that the dosage sensitivity of Wdr47 is also reflected at the behavioral level (Dataset S11). Together, these results support that the neuroanatomical defects associated with Wdr47 KO result in hyperactivity and sensory motor gating abnormalities both in male and female mice.
Fig. 6.
Assessment of behavioral traits in Wdr47 mouse models. Mice were analyzed for behavioral anomalies using 16 tests (Dataset S11). Here, we show a selection of results for two cohorts: one male (11 mice Wdr47+/tm1b vs. 11 mice WT) and one female (5 mice Wdr47tm1a/tm1a vs. 7 mice WT). (A) Traveled distance in centimeters and numbers of rears for circadian activity recorded for 32 h and open-field activity for a duration of 30 min. (B) Learning and memory were tested using the Y maze (short-term memory), Morris water maze (spatial memory), and novel object recognition with retention time of 24 h (long-term memory). (C) Skilled movements evaluated using the MoRaG. (D) Grip strength for both forelimb and hind limb. (E) Motor coordination assessed using the notch bar. (F) Pain sensitivity evaluated by the latency to react to heat. All plots are represented as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
Fig. S10.
Additional assessment of whole-body traits in Wdr47 mice. (A) Anxiety assessed using two tests: the elevated plus maze and the open-field test. (B) Ataxic-like movements recorded using the gait analysis. (C) Pain sensitivity evaluated using the shock threshold at which the mouse reacts by flinching, vocalization, or jumping. (D) Touch sensitivity evaluated using the adhesive removal test. (E) Social behavior studied using the social interaction test. (F) Social memory tested using the social recognition test. (G) Body weight measurements in male mice at eight different time points. (H) Weight of white adipose tissue and brown adipose tissue in KO mice compared with matched WTs. (I) Behavioral assessment in aged mice (56 wk old). Comparison between forelimb strength and hind paw errors at 16 and 56 wk of age mice using eight Wdr47+/tm1a vs. eight matched WT mice (male; same cohort evaluated at both ages). Number of mice used for A–H: female: n = 5 Wdr47tm1a/tm1a, n = 7 matched WT; male: n = 12 Wdr47+/tm1a, n = 5 matched WT. All plots are represented as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; #P < 0.07.
To test whether behavioral phenotypes worsen with age, we generated a 56-wk-old cohort but found no difference between phenotypes detected from the same cohort at 16 wk of age, suggesting that Wdr47 is unlikely to be implicated in neurodegeneration (Fig. S10I). In addition, we tested the impact of enriched diet on behavioral performances but saw no phenotypic improvement or rescue compared with mice on chow diet (Dataset S11).
WDR47 Plays a Role in Cell Homeostasis and Autophagy.
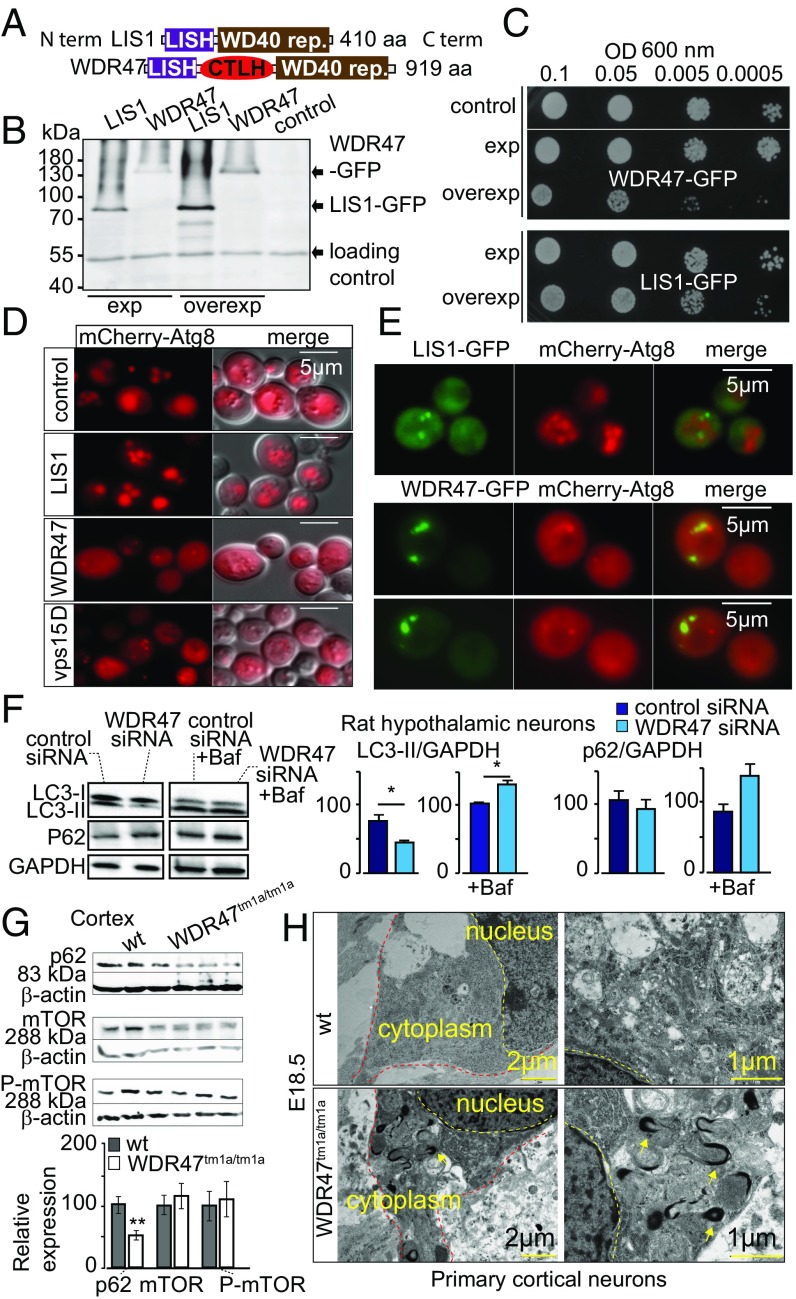
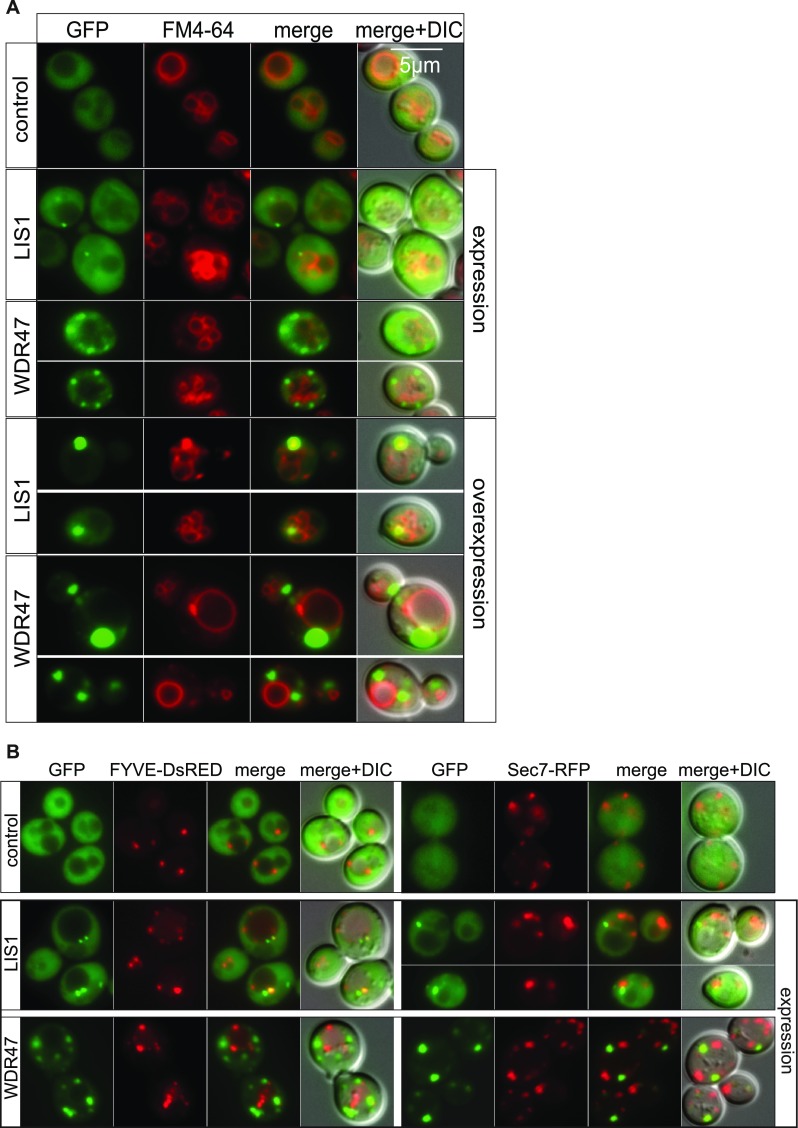
Several important WDR proteins implicated in autophagy [Atg18 (34), PIK3R4 (35)] have been identified in yeast before transposition to a mammalian system. WDR47 is not normally expressed in yeast; however, this unicellular model allows us to rapidly test for a wider spectrum of biological processes. We took advantage of this to study the WDR47 C-terminal to LisH (CTLH) domain, a predicted alpha-helical sequence with function that remains completely unknown in mammals (36) (Fig. 7A), by overexpression of human WDR47 and LIS1 in Saccharomyces cerevisiae (Fig. 7B). While WDR47 is less expressed than LIS1 (Fig. 7B), delayed growth was recorded exclusively in WDR47-GFP–transformed cells (Fig. 7C), suggesting that WDR47 is hijacking important cellular functions. After staining of the vacuolar membrane with the lipophilic dye FM4-64 (Fig. S11A), we observed that LIS1-GFP was mainly cytosolic; however, on overexpression, it associated with a large punctate structure adjacent to the vacuole, whereas WDR47-GFP was associated with smaller structures in the cytoplasm (Fig. S11A) but not with endosomes or Golgi complex (Fig. S11B). These results show that, when WDR47 and LIS1 are overexpressed in yeast cells, they have different intracellular localization, and only WDR47 overexpression impairs growth.
Fig. 7.
WDR47 is a key effector of autophagy. (A) WDR47 and LIS1 structures. (B) Western blot on yeast protein extracts with anti-GFP antibodies. (C) Drop test growth assays done on WT yeast cells (BY4742) transformed with pAG413 [low-copy number centromere (CEN) plasmid; expression] or pAG423 (2 microns; overexpression) plasmids bearing LIS1 or WDR47. Midlog phase cultures of the indicated yeast cells serially diluted to the indicated OD600 and spotted onto synthetic medium without histidine (SC-His). Growth evaluated after 2 d of incubation at 30 °C. (D) WT BY4742 (control) or vps15Δ (negative control) yeast cells transformed with mCherry-Atg8 plasmid and WT BY4742 cells cotransformed with expression plasmid (pAG413) bearing LIS1 or WDR47 cDNA observed by fluorescent microscopy after incubation for 4 h in nitrogen starvation medium (SD-N) to induce autophagy. (Scale bars: 5 μm.) (E) Living WT yeast cells (BY4742) expressing human LIS1-GFP or WDR47-GFP and mCherry-Atg8 observed by fluorescence microscopy after induction of autophagy by incubation in SD-N medium. (F) Western blot quantification of LC3 and p62 relative protein levels in the presence and absence of Bafilomycin A1 (Baf) treatment in response to WDR47 siRNA treatment. GAPDH was used as a loading control. (G) Western blot images of p62, mTOR, and phospho-mTOR in the cortex of WT and Wdr47tm1a/tm1a. Quantification of relative protein expression normalized against β-actin is plotted as mean ± SEM (n = 6 Wdr47tm1a/tm1a and n = 9 WT, male and female). *P < 0.05; **P < 0.01. (H) Transmission EM of cortical neurons from Wdr47tm1a/tm1a embryos (n = 3) at E18.5 compared with WT (n = 3). (Scale bars: Left, 2 μm; Right, 1 μm.)
Fig. S11.
WDR47 and LIS1 localization in yeast. (A) Living WT yeast cells (BY4742) expressing or overexpressing human LIS1-GFP or WDR47-GFP observed by fluorescence microscopy after staining of the vacuoles with the FM4-64 lipid dye. Merge shows the merge between the GFP and DsRED images, and Merge + DIC shows the merge between the GFP, DsRED, and DIC images. (B) Living WT yeast cells (BY4742) expressing human LIS1-GFP or WDR47-GFP observed by fluorescence microscopy to study localization in endosomal compartment positive for FYVE-DsRED (a probe for PtdIns3P that is localized on endosomes in yeast) and the Golgi complex (labeled with Sec7). (Magnification: 100×/1.45 oil objective.)
Autophagy being essential for cell viability on nutrient starvation in yeast (37) and autophagosomes being formed at a single site next to the vacuolar membrane (38), we asked whether WDR47 or LIS1 could be localized to autophagy sites. We first used expression of WDR47 or LIS1 with mCherry-Atg8, a homolog of mammalian LC3 (39). This revealed that WDR47 impaired yeast autophagy, whereas LIS1 did not. Indeed, mCherry-Atg8 did not reach the lumen of the vacuole and accumulated into the cytoplasm (Fig. 7D). Coexpression of WDR47- or LIS1-GFP with mCherry-Atg8 also impaired yeast autophagy and further showed that WDR47 was associated with punctate structures that did not colocalize with Atg8 (Fig. 7E). These results suggest that WDR47 might interact with some yeast autophagy effector, thereby inhibiting this cellular process.
To investigate this hypothesis in mammalian cells, we tested autophagy in GT1-7 WDR47 siRNA-treated cells and quantified levels of LC3 and p62 (or SQSTM1), two key proteins involved in autophagy. We found that LC3-II levels, but not LC3-I, were reduced (Fig. 7F). To assess whether this is caused by increased autophagic flux or decreased autophagosomal synthesis, we treated both WDR47 siRNA-treated and siRNA control groups with bafilomycin A1, a potent inhibitor of the H+ ATPase that reduces lysosomes functionality, and found that both LC3-II and p62 levels were increased (Fig. 7F), indicating enhanced autophagy flux. We also tested the expression of p62 and mTOR (an upstream regulator of autophagy) in cortices from adult Wdr47tm1a/tm1a, showing a clear specificity of WDR47 function in p62-mediated autophagy (Fig. 7G). In addition, transmission EM revealed abnormal autophagosomes in the cytoplasm of primary neuronal cultures (Fig. 7H). Together, these data show an additional role of WDR47 in cell homeostasis and protein clearance by modulating autophagic activity.
Discussion
WDR proteins have recently emerged in the field of neuroscience, but their function and ultimately, their participation in shaping the mammalian brain remain to be addressed. Here, we report the analysis of 26 WDR proteins in brain anatomy and focus on the functional characterization at the whole-organism level using a combination of cellular, invertebrate, and vertebrate model systems (siRNA, yeast, and KO mice) of a poorly characterized member, WDR47. WDR47 is also known as Nemitin (23), a protein sharing structural homology with LIS1, a WDR protein identified 20 y ago and associated with lissencephaly (7). Our findings highlight four important points.
First, 27% of assessed WDR genes gave rise to severe brain anomalies when inactivated in mice, showing the functional importance of WDR genes in brain connectivity, particularly in the genesis of the corpus callosum, a commissure that provides higher-order neurological advantages in placental mammals. This is in line with the emerging role of WDR genes in human brain pathologies associated with corpus callosum anomalies. Examples include WDR73, WDR81, ERCC8, and HERC1 (40). Herc1−/− was coincidently processed in our study and showed macrocephaly and enlarged corpus callosum, reminiscent of the radiographic features of human patients with HERC1 mutations, showing the pertinence of rodent screens for translating neuroanatomical disorders in humans.
Second, while WDR47 has previously been shown to associate with microtubules (23), we report here its implication in brain development and corpus callosum genesis. Our working model is shown in Fig. 8. In the absence of WDR47, we show that both callosal and corticofugal neurons have severe fiber tract defects and abnormally shaped growth cones in conjunction with microcephaly linked to the exhaustion of late cortical progenitors and the consequent decrease of neurogenesis, possibly yielding to abnormal genesis of glial cells. Together, these could provide a molecular model underlying altered motor coordination, skilled movements, and pain sensitivity. Our work also provides insights showing that WDR47 plays a microtubule stabilizer role in the growth cone. This is further supported by the interaction between the N terminus of WDR47 and SCG10 [a very well-known microtubule destabilizer promoting catastrophe at the growth cone (41)], which led us to think that WDR47 might also be a regulator of SCG10 activity in the JNK1 pathway. Interestingly, KO mouse studies of Jnk1 (42) show high behavioral similarities (for example, motor coordination defects) compared with Wdr47.
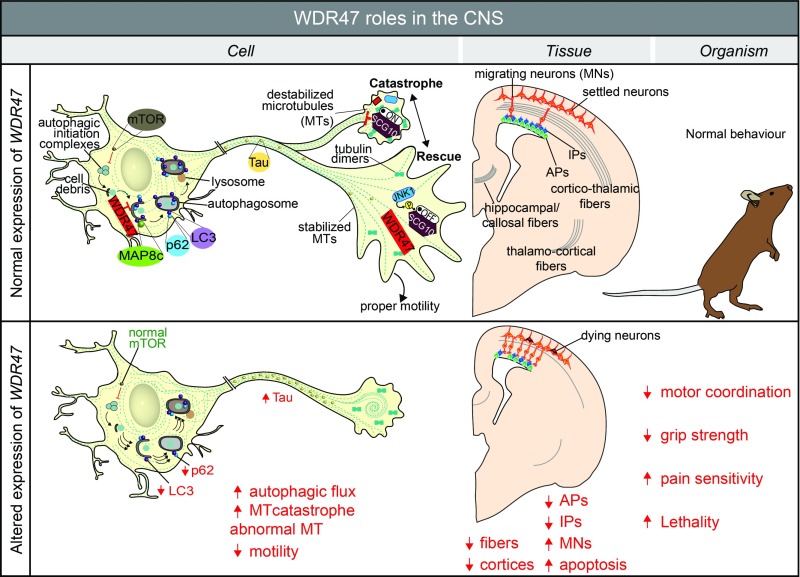
Fig. 8.
Summary of WDR47 working model. At the cellular level, WDR47 acts as a stabilizer of microtubules. SCG10, a microtubule destabilizer at the growth cone, is regulated by JNK1 through phosphorylation. In the cytoplasm, WDR47 negatively regulates p62 and LC3-mediated autophagy. Reduced expression of Wdr47 leads to destabilization of the microtubules, increased Tau and autophagic flux, and decreased cell motility. At the tissue level, the loss of Wdr47 yields fiber tract defects, including corpus callosum agenesis, microcephaly, and thinner cortices, that are linked to a reduced number of progenitors, increased number of neurons in the intermediate zone, and elevated number of dying neurons in the cortical upper layers. At the organism level, these defects may explain impairment in motor coordination, reduced grip strength, and increased pain sensitivity. AP, apical progenitor; IP, intermediate progenitor; MN, migrating neuron; MT, microtubule.
Third, altered levels of LC3-II and p62 proteins, concomitantly with increased autophagy flux, suggest that WDR47, unlike LIS1, is involved in the regulation of key effectors of autophagy in the brain. Autophagy is a protein clearance process of aberrant or obsolete cellular structures and organelles crucial in cell homeostasis. Microtubules seem an important player in the autophagy process in mammals (43); however, their specific implication in autophagy is unclear (44). A potential molecular mechanism by which WDR47 regulates autophagy might be through its interaction with the light chain of MAP8 (also known as MAP1S) (23). In a similar way to MAP8 (45), WDR47 might bridge the autophagy machinery, in particular, LC3-II–bound autophagosomes, with microtubules. Unlike other cells of the mammalian system, postmitotic neurons are more vulnerable to damage from cellular debris and thus, would require a well-regulated degradation pathway. WDR47 involvement in the survival of postmitotic neurons in late corticogenesis from upper cortical layers, where WDR47 is the most expressed, could be mediated through the proper regulation of autophagy, reinforcing the molecular cross-talk between autophagy and apoptosis (46). Furthermore, the possible association of WDR47-related autophagy to brain wiring is supported by a recent report showing that the WDR autophagy scaffold protein (ALFY) is required for neuronal connectivity in the mouse brain (47). Strikingly, the lack of Alfy in mice led to perinatal lethality, microcephaly, absence of the corpus callosum, and hypoplasia of the internal capsule (47), reminiscent of Wdr47 phenotypes, suggesting a similar mode of action. It may, therefore, be that WDR47 regulates the cell ability to detect environmental cues via its microtubule stabilization role, which in turn, regulates the protein degradation pathway necessary for neuronal shape and motility.
Fourth, the final significant finding is the essential role of WDR47 for survival. The scarcity of patients harboring mutations in WDR47 supports this notion. Indeed, we found no truncating mutations in WDR47, despite sharing data at numerous genetic meetings, and only three stop-gained mutations are reported in the ExAC database (48), suggesting a selection bias against LoF mutations in WDR47. Extending our analysis to three specific cohorts of unknown genetic cause made up of patients with lissencephaly, intellectual disability, and nonsyndromic agenesis of the corpus callosum, we identified three missense variants (Datasets S12 and S13); however, pathogenicity and transmission mode were not compatible with WDR47 gene causality. The inability of S. cerevisiae to grow, possibly because of the inhibition of autophagy on overexpression of WDR47, further supports the essentiality of WDR47 for survival but also suggests that mirrored protein levels (too little or too much) have identical effects on pathogenicity and underlying biological processes. This is a characteristic of scaffold proteins, indicating that WDR47 might be serving as a support for the interaction of other proteins, such as SCG10. Although we were unable to identify the precise reason of death, cardiac and breathing failure were excluded. Autophagy being dramatically up-regulated in the neonatal stage to overcome the starvation period (49), mice could be dying of lack of nutrients, since an enriched lipid diet rescued lethality.
In conclusion, this study presents identification of the relevance of WDR genes in brain connectivity, highlighting the power of unbiased and high-throughput mouse LoF studies in the field of neuroscience. These mouse models could explain some of the missing genetics in corpus callosum biology (11) and help pave the way for a better stratification system of complex neurodevelopmental steps. WDR47 plays a role in the regulation of microtubule dynamics, progenitor proliferation, neuronal migration, and fiber tract projections in a similar fashion to LIS1 (50, 51) but with the distinctive particularity that WDR47 inhibits autophagic flux. This provides a functional link between autophagy biology and the CTLH domain (52) in mammals [this association was previously made with the vacuole import and degradation pathways in yeast (53)], while strengthening the emerging link between autophagy and microtubules assembly. Although a definite association of WDR47 with human brain disorders has not been made yet, WDR47 should be considered as a candidate gene for corpus callosum abnormalities and motor coordination deficiencies, possibly through compound heterozygosity or somatic mosaicism. Considering that the microtubule stabilizer drug restored growth cone defects, it will be interesting to test whether the behavioral anomalies, as already shown in an Alzheimer mouse model (54), could also be restored. This might open up therapeutic perspectives in the clinic with the aim of relieving symptoms in patients suffering from this class of diseases that we refer to as “WDRopathies.”
Materials and Methods
Mutant mice were obtained through collaboration with the Sanger Mouse Genetics Project. Wdr47-targetted mice were generated using the International Mouse Phenotyping Consortium targeting mutation strategy (55). This relies on the identification of an exon common to all transcript variants, upstream of which an LacZ cassette was inserted to make a KO (tm1a), whereas tm1b creates a frameshift mutation on deletion of the selected exon. Animal procedures were approved by the local ethics committee (Com’Eth) under the reference number 2016010717527861. DNA samples from patients and their parents and informed consent were obtained in accordance with the local ethics committee of the Strasbourg University Hospital (Comité Consultatif de Protection des Personnes dans la Recherche Biomédicale). Full methods are described in SI Materials and Methods.
SI Materials and Methods
Analysis of WDR Genes.
We used SMART (simple modular architecture research tool) protein database to retrieve any proteins associated with a domain encompassing the term “WD40-repeat-containing” with one-to-one orthologs between human and mouse genomes. We manually curated this list, verified from the literature whether the protein had synonyms and whether it was associated with a human disease using MGI (www.informatics.jax.org/), and finally, removed any noncoding proteins. This produced a list of 286 unique WDR proteins, common to both species (Dataset S1). We carried out ontology enrichment analysis using ToppGene (https://toppgene.cchmc.org/). A total of 181 brain measurements distributed in two coronal histological sections at Bregma +0.98 and −1.34 mm were taken with a precision down to cell-level resolution (Dataset S2) in conjunction with 14 quality control parameters (Dataset S3). When applicable, we averaged measurements from left and right hemispheres. To avoid any biases, the same experimenter carried out all brain analyses of 26 WDR genes, and this was done completely blind to the genotype. To determine whether a gene is associated with morphological anomalies, we utilized Phenstat, a software developed for high-throughput phenotyping programs, such as the International Mouse Phenotyping Consortium (56), with a high confidence significance threshold of less than 0.0001 accounting for multiple testing. Statistical analyses were done using R Studio (version 3.3.1). The use of C57BL/6N as genetic background is highly relevant, since it is clear of corpus callosum anomalies unlike other strains (57).
Animal Welfare.
The mice were bred at the Mouse Clinical Institute under controlled light/dark cycles and were provided with food and water ad libitum. We maintained two independent colonies; each was fed with a different diet: the first with chow diet (D04; Safe Diets Laboratory) and the second with an enriched diet (DIET-5021–3-BG; Autoclavable Mouse Breeder Diet 5021). All animal procedures were carried out in accordance with the ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines and approved by the animal ethics committee under number 2016–017.
Wdr47 KO Construction.
The construction of alleles is based on the “Knockout-first allele” method (55) (Fig. S1B). A “critical” exon (exon 5) present on all transcripts was first identified. A promotorless targeting cassette, ES cell clone EPD0046_1_C04 from Knockout Mouse Program, was inserted in intron 4 in C57BL/6N blastocyst, which created a frameshift mutation, thus generating the KO-first allele (tm1a allele); tm1a mice were bred into mice expressing Cre recombinase under the ROSA26 promoter to generate null alleles (tm1b) and into mice expressing Flp recombinase germ-line deleter mouse to generate floxed animals (tm1c). A het-by-het breeding scheme was used to propagate the various lines.
Wdr47 Genotyping.
Genomic DNA was extracted from mouse ear or tail clippings. Primers were designed specifically for each mouse model (Fig. S1B) with the following sequences: Ef 5′AGGTTGTCATGCAGTCTGGG3′, Er 5′GGATGACTATAAAGCGGTGCAAG3′, Kr 5′CTCCTACATAGTTGGCAGTGTTTGGG3′, L3f 5′TCCTTTGCTAACTTCCACTATCC3′, L3r 5′TCAGCCTGGTCTACAGAGTTA3′, Lxr 5′ACTGATGGCGAGCTCAGACCATAAC3′, and Mq1f 5′GGGATCTCATGCTGGAGTTCTTCG3′. PCR reactions were carried out using the FastStart PCR Master (Roche Life Science) under the following thermocycler conditions: 95 °C for 4 min (1 cycle), 94 °C for 30 s followed by 62 °C for 30 s and 72 °C for 1 min (34 cycles), 72 °C for 7 min (1 cycle), and 20 °C for 5 min (1 cycle). Expected size of PCR product is shown in Fig. S1B.
Touch Down PCR.
When PCR did not work, we used a touch down system instead. The first step consisted of enzymatic activation at 94 °C for 5 min (1 cycle); for 13 cycles, 94 °C for 30 s (DNA denaturation), 57 °C for 30 s (primers annealing temperature was programmed, so that 0.5 °C was decreased after each cycle, resulting in 51 °C at the 13th cycle), and 72 °C for 45 s (primers extension); and for 29 additional cycles, 94 °C for 30 s, 51 °C for 30 s, 72 °C for 45 s, and a final cycle at 72 °C for 5 min.
LacZ Expression.
Brain samples from mice at E14.5, E16.5, E18.5, and 16 wk of age and whole embryos at E9.5 were collected. Samples were dropped–fixed in fixative solution (10% formaldehyde, 0.4% gluteraldehyde, 0.04% Np40, 0.02% NaDC) for 2 h and washed in 1× PBS. The samples were incubated in X-Gal solution (5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 1 mg/mL X-Gal, 2 mM magnesium chloride in 1× PBS) at 4 °C overnight. The samples were washed in 1× PBS, sectioned at 50-µm thickness using a vibratome, and postfixed in 4% paraformaldehyde (PFA). The sections were counterstained in 0.1% Safranin solution and mounted on slides before imaging using the Hamamatsu slide scanner.
Wdr47 qPCR.
Total RNA was extracted from brain and peripheral regions (olfactory bulb, cerebellum, medulla, cortex, hippocampus, thalamus, striatum, spinal cord, heart, lung, and liver) using a phenol-chloroform technique and RNeasy Plus Mini Kit (74134; Qiagen); 1 µg of total RNA was reverse-transcribed to cDNA using SupercriptIII First-Strand Synthesis Supermix for qRT-PCR (11752–050; Invitrogen). PCR reactions were performed using Fast Blue qPCR Mastermix Plus (RT-QP2X-03+FB; Eurogentec). Primers were designed as 5′GGACCCCAGTGGCCGTCTCT3′ (forward), 5′GCTCTTTCTGGGGCAGGACGC3′ (reverse), and 5′TGATCCTTCAGAAGCAGCCA3′ (probe). The samples were run in triplicate and normalized against housekeeping gene GNAS (5′AGAACATCCGCCGTGTCTTC3′, 5′CCTTCTTAGAGCAGCTCGTATTGG3′, and probe 5′CGTGACATCATCCAGCGCATGCAT3′). Gene expression was analyzed using the delta cycle threshold method (58).
Viability Test.
At E9.5, 34 fetuses were isolated from their embryonic sac after caesarean delivery. The uterine membrane was carefully removed, thus separating the placenta from the yolk sac. The live embryo within the yolk sac was kept in 1× PBS and observed under the stereomicroscope for abnormalities in number of somites, limb morphology, heartbeat, and blood circulation. The embryo was carefully dissected out of its yolk sac, fixed in 4% PFA for 24 h, transferred to 70% ethanol, and stored at 4 °C. The yolk sac was used for genotyping. At E18.5, 56 fetuses were isolated from six pregnant females through caesarean delivery. The fetuses were placed on a heating plate maintained at 37 °C and rolled gently to stimulate breathing. They were observed for 30 min after delivery. We quantified the number of embryos breathing properly, moving, and turning pink as opposed to cyanotic animals that were unable to breathe. Mice that survived were euthanized with an i.p. injection of 50 μL anesthetic (750 µL rompun, 1 mL ketamine, 4 mL 0.9% NaCl). Tail samples were harvested for genotyping. We also assessed 11 cohorts of mice right after normal delivery. We studied the mice thrice every day (4-h intervals) using a set of parameters, including general health status, skin color, movements, breathing capacity, brain morphology, weight, and physical size. When a mouse died, time of death was recorded along with other parameters (weight, head circumference, and length of body), and the tail was collected for genotyping. The mice were observed until weaning age.
Western Blot Analysis.
The cortices of adult and E12, E14, E16, and E18 embryonic mice were dissected and homogenized with 300 μL of lysis buffer containing 1× RIPA buffer (ThermoFischer), phenylmethylsulfonyl fluoride 1%, sodium orthovanadate 1%, and protease inhibitor 1% in tubes containing ceramic beads (Precellys Lysing Kit). The tubes were incubated for 30 min at 4 °C and centrifuged for 20 min at 17,000 × g at 4 °C, and the supernatant was isolated for Western blotting; 50–55 μg of protein was separated on 12% or 6% SDS/PAGE depending on the target protein size and transferred onto nitrocellulose membrane (BioRad). Membranes were blocked with 5% nonfat dry milk diluted in tris buffered saline with Tween 20 (50 mM Tris, 150 mM NaCl, 0.05% Tween 20) and probed using the antibodies: rabbit polyclonal anti-WDR47 (ab121935; 1:50 dilution; Abcam), rabbit polyclonal mTOR (SAB4501038; 1:500 dilution; Sigma-Aldrich), rabbit polyclonal p-mTOR (SAB4504476; 1:1,000 dilution; Sigma-Aldrich), guinea pig polyclonal anti-p62 (GP62-C; 1:200 dilution; Interchim SA), and rabbit polyclonal to Tau (nonphosphorylated and phosphorylated at S262; ab64193; 1:200 dilution; Abcam) overnight at 4 °C. β-actin (A3854; 1:50,000; Sigma-Aldrich) and GAPDH (G9545; 1:1,000 dilution; Sigma-Aldrich) were used as loading controls. Antibody–protein interactions were revealed using chemiluminesence (RPN2108; GE Healthcare), and relative protein expression was quantified using ImageJ (https://imagej.nih.gov/ij).
Histology.
Adult mice were euthanized in a CO2 chamber, and brains were dissected and drop fixed in 10% formalin. The samples were transferred to 70% ethanol 48 h after drop fixation. They were trimmed either at the midline on sagittal plane to produce two hemispheres or at two defined coronal planes to produce three blocks (the latter is explained in detail in ref. 59) and embedded in paraffin using an automated embedding machine: Sakura Tissue-Tek VIP. In the case of embryos at E18.5, the whole embryo was fixed in Bouin solution for 7 d. The brains and lungs were extracted, transferred to 70% ethanol, and manually embedded in paraffin using the following steps: three incubation baths in 70% ethanol for 30 min each, two baths in 95% ethanol for 30 min each, two baths in 100% ethanol for 45 min each, three baths in Histosol Plus for 1 h each, and five baths in warm paraffin for 30 min each followed by incubation in paraffin overnight before casting in a mold. Brains were cut at a thickness of 5 µm on a microtome, such that we obtain sections matching planes displayed in Fig. S2 for sagittal sections and Fig. S5 for embryonic coronal sections. The sections were then stained with 0.1% Luxol Fast Blue (Solvent Blue 38; Sigma-Aldrich) and 0.1% Cresyl violet acetate (Sigma-Aldrich) and scanned using Nanozommer 2.0HT, C9600 series at 20× resolution. Embryonic lung sections were also cut at 5 µm on a microtome, stained using H&E, and observed for morphological defects.
Histomorphometric and Statistical Analyses.
Each image was quality controlled to assess whether (i) the section is at the correct position, (ii) the section is symmetrical, (iii) the staining is of good quality, and (iv) the image is good quality. Only images that fulfilled all of the quality control checks were processed. To avoid experimenter bias, the same person measured the 78 parameters on adult coronal planes, 67 parameters on embryonic planes, and 95 parameters on sagittal planes (Dataset S8). We also checked for the presence of erroneous measures, typographical errors, and outliers and corrected the measurement if necessary. We used a linear mixed model as implemented in Phenstat (56) to assess significance and critically evaluated any results with P values higher than 0.0001. We considered a phenotype hit if sections were undamaged and if measurements were accurate.
Immunohistochemistry.
Sample processing.
Embryonic brains were dissected in 0.1 M PBS, pH 7.4 and were fixed at 4 °C in 4% PFA overnight. For cryosections, fixed samples were cryoprotected overnight in 20% sucrose in PBS at 4 °C, embedded in OCT compound (Sakura Finetek Europe B.V.), and either sectioned at 16–18 µm onto slides (SuperFrost Plus; VWR International) using a cryostat (CM30505; Leica Biosystems) or sectioned at 50-µm thickness using a frozen sliding microtome (HM450; Microm). For vibratome sections, fixed samples were washed in PBS, embedded in 4% low-melt agarose, and sectioned (60 µm) using a vibratome (VT1000S; Leica Biosystems).
Immunostainings.
For staining, both cryostat and vibratome sections were rinsed in PBS, blocked in 10% Normal Donkey Serum/PBST (phosphate buffered saline with Triton; 1× PBS, 0.3% Triton X-100), and incubated with primary antibodies at 4 °C overnight. After washing with PBST, the sections were incubated for 1.5 h at room temperature with fluorescence-conjugated secondary antibodies coupled to Alexa-488, Alexa-647, or Alexa-555 (Thermo Fischer Scientific). Nuclei were counterstained with DAPI (1:10,000 dilution; Sigma-Aldrich), and sections were mounted using Aquapolymount mounting solution (Polysciences Inc.). The slides were stored in the dark at 4 °C. The following antibodies were used: rabbit polyclonal antiactivated caspase-3 (AF835; 1:100 dilution; R and D Systems), mouse monoclonal anti-Ki67 (clone B56; 1:500 dilution; BD Pharmingen), rabbit polyclonal Ki67 (IHC-00375; 1:500 dilution; Bethyl Laboratories), rabbit polyclonal anti-Pax6 (AB2237; 1:500 dilution; Merck Millipore), rat monoclonal anti-Tbr2 (14–4875-80; 1:250 dilution; eBioscience), chicken anti-GFP (GFP-1020; 1:800 dilution; Aves Labs Inc.), mouse antineurofilament panaxonal antibody (SMI-312R; Covance and 837904; 1:1,000 dilution; Biolegend), and rat antineural cell adhesion molecule L1, clone 324 (MAB5272; 1:250 dilution; Merck Millipore).
EdU labeling and detection.
To examine cell cycle exit and fate of newborn cells, EdU (41 µg/mg) was injected i.p. into pregnant mice at E15.5, and embryonic brains were collected 24 h later at E16.5. To analyze positioning of neurons, EdU was injected at E14.5, and embryonic brains were collected 4 d later at E18.5. After fixation and cryoprotection, frozen sections of embryonic brains were processed for EdU detection with the Click-iT EdU Alexa Fluor 647 kit (Thermo Fischer Scientific) according to the manufacturer’s protocol.
Image analysis, cell counting, and statistics.
Images were acquired using confocal microscope (TCS SP5; Leica) at 20× magnification and analyzed using ImageJ software. Cortical wall areas were identified according to cell density and nuclear orientation (nuclei staining with DAPI). The total number of markers-positive cells in the embryonic brain sections was quantified by counting positive cells in a rectangle of 100-µm width with anatomically matched positions in experimental groups. Results were obtained from at least three mouse brains of each genotype. For migration experiments with EdU and in utero electroporation data analysis, the numbers of marker-positive cells were counted, and the percentage of positive cells in each cortical area (ventricular zone/subventricular zone, intermediate zone, and cortical plate) was calculated. For SMI-312R and L1CAM experiments, stained sections were scanned using the Nanozommer 2.0HT, C9600 series at 20× resolution and imaged at higher resolution using a confocal SP5 inverted microscope at 40× or 63× oil lens and a z stack of 0.1 μm. Statistics for dual comparisons were generated using unpaired two-tailed Student’s t tests, while statistics for multiple comparisons were generated using two-way ANOVA followed by appropriate post hoc test (version 6; GraphPad Prism software) with *P < 0.05, **P < 0.01, and ***P < 0.001 for all statistics.
In Utero Electroporation.
Timed pregnant mice (E14.5) were anesthetized by isoflurane and put on a heated pad. The uterine horn was exposed, and 0.5–1 μL of Endofree plasmid DNA solution containing 1 μg/μL of NeuroD:Cre-GFP and 2 μg/μL of NeuroD:GFP was mixed with 0.05% Fast Green (Sigma-Aldrich) and injected into the lateral ventricle of the embryonic brain with a fine glass micropipette and Femtojet microinjector (VWR International). Embryos were then clamped between 5-mm platinum tweezer electrodes, and five 50-ms, 35-V electrical pulses at an interval of 950 ms were applied using an ECM-830 BTX square wave electroporator (VWR International). Uterine horns were then placed back into the abdominal cavity, and the abdomen wall and skin were sutured using surgical needle and thread. The pregnant mouse was injected with metacam and warmed on heating pad until it woke up. Four days after surgery, pregnant mice were killed by neck dislocation, and embryos were processed as described above.
Immunocytochemistry and Epothilone Treatment in Primary Neuronal Cells.
Dissociated mouse hippocampal and cortical neurons from embryos dissected at E17.5 were cultured in GIBCO Neurobasal Medium 1× (Invitrogen) supplemented with GIBCO B27 Medium (Invitrogen) and l-glutamine and sustained at 37 °C with 5% CO2. For immunocytochemistry, cells were cultured on poly-l-lysine–coated 24-well plates. The cells were fixed at 4 d in vitro with 4% PFA in 6% sucrose. The cells were stained using a range of antibodies: rabbit polyclonal anti-WDR47 (ab121935; 1:50 dilution; Abcam); rabbit polyclonal anti-MAP2 (AB5622; 1:1,000 dilution; Millipore), which stains the cell body and dendrites; mouse monoclonal antineurofilament panaxonal antibody (SMI-312R; Covance and 837904; 1:1,000 dilution; Biolegend), which stains the axon and growth cone; mouse monoclonal anti-STMN2 (MABN663; 1:50 dilution; Merck Millipore); goat polyclonal anti-JNK1 (sc-46006; 1:50 dilution; Santa Cruz Biotechnology); and mouse monoclonal antiacetylated tubulin clone 6–11b-1 (T6793; 1:200 dilution; Sigma-Aldrich), a marker for stabilized microtubules. Images were taken using a regular fluorescence microscope (Leica). Length and area measures were made on cells stained with anti-MAP2 and SMI312 using ImageJ. Axonal length was analyzed by measuring the length from the emergence of the axon at the cell body to the tip of the primary axonal process, stopping when the growth cone begins.
Neuronal cultures were treated with 10 and 100 nM EpoD, a microtubule-stabilizing drug dissolved in DMSO for 1.5 h before being fixed in a 4% PFA and 6% sucrose solution. The cells were then stained with SMI312 to study growth cone morphology. For live imaging, cells cultured on Labtek I four-well culture chambers (Nunc) coated with poly-l-lysine and laminin were maintained in optimum conditions (37 °C, 5% CO2) and recorded in DIC (differential interference contrast) imaging settings on inverted microscope (Leica) using MetaMorph (Molecular Devices).
MRI.
Sample preparation.
Wdr47tm1a/tm1a and WT mice, age 16 wk old, were anesthetized i.p. with 0.1 mL/10 mg of body weight ketamine (150 mg/kg)-xylazine (10 mg/kg) in saline solution. They were perfused with 30 mL of 1× PBS solution containing 0.4% ProHance Gadoteridol (Bracco Imaging) and 10 mg/100 mL heparin followed by 30 mL of 4% PFA-0.4% ProHance at a rate of 1 mL/min. The brains remained in the skull with the skin trimmed off and were maintained in 4% PFA-0.4% ProHance overnight at 4 °C. The specimens were transferred into 1× PBS-4% ProHance the following day and kept at 4 °C until imaging.
Diffusion tensor protocol.
MRI experiments were conducted on a 7-T Biospec 70/30 USR NMR spectrometer (Bruker Biospin) at 20 °C operating under PARAVISION 6.01 software (Bruker BioSpin). Transmission was achieved with a quadrature volume resonator (i.d. 86 mm), while a mouse brain quadrature surface coil (19 × 19 mm2) was used for signal reception (Bruker BioSpin).
Diffusion tensor data acquisition.
Data were acquired using a diffusion tensor spin-echo echo-planar imaging sequence with the following parameters: three shots, 20 averages, TR/TE (repetition times over echo time) = 1,680/30.5 ms, 20 consecutive slices with resolution = 0.1 × 0.1 × 0.3 mm, ∆/δ = 11.2/5.5 ms, 30 directions, four b values = 0, 650, 1,000, and 2,000 s/mm2. Scan time was 2 h and 33 min.
Image postprocessing.
Motion corrections were applied by individual coregistration using the b-value null as reference (FNIRT FMRIB Software Library). Diffusion tensor data were analyzed using DSI Studio (dsi-studio.labsolver.org) to obtain the main direction of the tissue represented by color maps combining FA (fractional anisotropy), directional map, and tractography. Tractography was performed on whole brain based on a streamline tracking method with the computation terminated after 100,000 seeds using the following parameters: FA threshold = 0.25, turning angle = 65.
Transmission EM.
Primary neurons at E18.5 were fixed in 2.5% glutaraldehyde-2.5% PFA in cacodylate buffer (0.1 M, pH 7.4) and then washed in cacodylate buffer for 30 min. This was followed by a postfixation step in 1% osmium tetroxide in 0.1 M cacodylate buffer for 1 h at 4 °C and staining with 2% uranyl acetate for 1 h at 4 °C. The neurons were then washed in 50, 70, 90, and 100% ethanol and propylene oxide for 30 min each. The neurons were then embedded in Epon 812, and semithin sections at 2 μm and ultrathin sections at 70 nm were cut and contrasted with uranyl acetate and lead citrate. The neurons were then examined at 70 kV with a Morgagni 268D electron microscope, and images were captured digitally by Mega View III camera (Soft Imaging System).
Superresolution Single-Molecule Localization Microscopy.
Superresolution single-molecule localization (SML) images were acquired using the Bruker Vutara 352 superresolution microscope. Acetylated tubulin labeled with Alexa-647 was imaged using a 60× N.A. 1.2 water objective (Olympus) and was excited using a 639-nm laser. SML superresolution data were reconstructed and processed using Vutara SRX software (Bruker).
Migration Assay and Imaging of GT1-7 Cells.
Rat GT1-7 hypothalamus-derived neuronal cells were cultured in DMEM supplemented with 10% FCS and 1% PenStrep. Cells were transfected for 24 h with HiPerfect Transfection reagent (Qiagen) and 28 ng/µL (12 µL) of WDR47 siRNA or negative control siRNA (Qiagen). An in vitro neuronal migration assay was performed by creating a linear wound in cell culture dishes, treating cells with 10 µg/mL of Mitomycin C (Sigma-Aldrich) to inhibit neuronal proliferation, and capturing images of the wound every 6 h for 36 h using an Olympus Cell^R system attached to an IX 81 inverted fluorescence microscope equipped with an F-view-II–cooled CCD camera (Soft Imaging Systems). Images were acquired using a Halogen light source and a transmission filter. Migration distances (perpendicular distance in micrometers from the wound edge) and wound areas were determined using Cell^R software. The percentage of wound closure was determined from wound area measurements (28).
Superresolution structured illumination microscopy (SR-SIM) was performed on transfected or fixed cells immunostained with mouse acetylated tubulin primary antibody (Santa Cruz) and Alexa Fluor 568 donkey anti-mouse IgG secondary antibody (Life Technology). Thin (0.1-μm) z stacks of high-resolution image frames were collected in five rotations by utilizing an alpha Plan-Apochromat 100×/1.46 oil DIC M27 ELYRA objective using an ELYRA S.1 (Carl Zeiss Microimaging) microscope equipped with a 488-nm laser (100 mW), a 561-nm laser (100 mW), and an Andor EM-CCD camera (iXon DU 885). Image analysis was performed on reconstructed superresolution images using ZEN software. To assess neuronal surface morphology, migrating neurons were fixed onto cover glasses 24 h postscratch introduction, double coated with gold to make the surface electrically conducting, and subjected to scanning EM. To assess neuronal ultrastructure, fixed cell pellets were processed and subjected to transmission EM.
Behavioral Analysis.
Behavioral experiments were conducted at the Mouse Clinical Institute (Illkirch, France) by the same experimenter blinded to the genotype to avoid any potential biases. All mice tested were given several days of rest between experiments and were age matched. Four independent groups of mice were used for behavioral experimentation, including three male cohorts (11 mice Wdr47+/tm1b vs. 11 matched WT, 8 mice Wdr47+/tm1a vs. 8 matched WT, and 12 mice Wdr47+/tm1a vs. 5 WT) and one female cohort (5 mice Wdr47tm1a/tm1a vs. 7 matched WT) (Dataset S11). We used a comprehensive pipeline of 16 behavioral tests, including (i) circadian activity for 32 h, (ii) open field, (iii) elevated plus maze, (iv) Y maze, (v) Morris water maze, (vi) novel object recognition 3 h, (vii) novel object recognition 24 h, (viii) MoRaG (32), (ix) grip strength, (x) notch bar, (xi) DigiGait, (xii) hot plate test, (xiii) adhesive removal test (33), (xiv) shock test, (xv) social interaction, and (xvi) social recognition.
Circadian activity.
The circadian activity test is used to assess spontaneous activity and feeding across the entire light/dark cycle. The mice were put in individual cages (11 × 21 × 18 cm3) fitted with IR captors linked to an electronic interface (Imetronic), which recorded locomotor activity for a period of 32 h; this included the habituation phase of 8 h followed by testing for 12 h each in the dark and light phases. Feeding behavior was also evaluated using a lickometer and pellet feeder (test Diet; Hoffman La-Roche).
Open field.
The open-field test encompasses studying basic locomotor activity, hyperactivity, exploratory behavior, and anxiety in mice. The mice were put in an arena of dimensions 44.3 × 44.3 × 16.8 cm made of PVC (Panlab) fitted with two frames of 32 IR beams and illuminated at 150 Lux, and their activity was recorded for 30 min using a video tracking system (Ethovision; Noldus); data were analyzed using Acti-Track software. Speed and distance covered by the mice were then quantified as well as the percentage of time spent in defined zones of the field (periphery, intermediate, or center).
Elevated plus maze.
An additional test was used to evaluate anxiety, the elevated plus maze (Imetronic). The setup consists of a plus-shaped arena illuminated at 50 Lux at an elevation of 66 cm, with opposite arms being either open (without walls 30 × 5 cm) or closed (with walls 30 × 5 × 15 cm). The apparatus is equipped with IR captors, allowing the detection of the mouse in the enclosed arms and different areas of the opens arms. The mouse was placed in the center and allowed to freely explore the arms for a period of 5 min. The index of anxiety is calculated against the number of entries and time spent in the closed arms as opposed to open arms.
Y maze.
The Y-maze test was used to evaluate short-term working memory based on the innate curiosity of the mice to explore an arm that has not been previously explored; this preferential behavior, when occurring at a frequency greater than 50%, is called spontaneous alternation. The testing apparatus consists of a Y-shaped maze with three white, opaque, Plexiglas arms of equal length (40 × 9 × 16 cm) at 120° angles to each other, each identified with walls designed with unique motifs, illuminated at 100 Lux. Animals were placed at the center of the maze and allowed to freely explore the three arms for 8 min. The numbers of arm entries were recorded and quantified to yield percentage of alternation.
Morris water maze.
The Morris water maze was used to evaluate spatial learning and memory. The water maze consists of a circular pool (radius 75 cm, height 60 cm) that is filled with 40 cm of water, which is made opaque using an aqueous acrylic emulsion polymer, maintained at 22 °C, and illuminated at 100 Lux. It is surrounded by various visual cues and also contains a platform of 5-cm radius submerged 1 cm below the water surface. The test spanned across 5 d of habituation and two tests (one on the last day of habituation and another 48 h later). During each habituation session, the mouse was placed in the pool facing the interior wall, and time taken to find the platform, velocity, and distance traveled were recorded; the maximum searching period allotted was 120 s. The platform remained constant throughout the tests; however, the mouse was placed in different quadrants each time. For the test sessions, the platform was removed, and the same parameters as well as the number of times that the mice crossed the quadrant and the exact position where the platform was previously kept were evaluated. We performed an additional test where the external visual cues were removed; however, a flag is placed on the platform for the mice to see so as to control their vision.
Novel object recognition.
The novel object recognition is similar to the Y-maze test, as it is based on the innate tendency of mice to explore novel objects over familiar ones. The animals were habituated (100-Lux illumination) and tested (70-Lux illumination) in the open-field arena. They were first presented with object A, a glass marble or plastic dice, and the exploration time was recorded. After a retention period (typically 3 h to test short-term memory or 24 h to test longer-term memory), the mice were presented again with object A together with a novel object B, and the exploration time of both the objects was recorded. The calculation was based on the percentage of time spent exploring the new object over total time spent exploring both objects.
MoRaG.
The MoRaG test was used to study forelimb reaching and grasping as well as laterality in forelimb movement of the mice (32). The mice were kept under restricted food allowance from the previous day to maintain 80–85% of their body weight. The testing apparatus consists of four chambers (one for testing and three for habituation) large enough to house a mouse that are made of clear Plexiglas (illuminated at 120 Lux), each fitted with a tiny circular window for the mouse to extend its paw through and a small platform outside the window to support a food pellet. The test spanned across 5 d (2 d of habituation and 3 d of test); 30 (for the first day) and 50 (for the remaining days) single food pellets (20 ng) were continuously presented to the mice, and the mice were scored for (i) their preferred paw to retrieve the pellet, (ii) their ability to reach the pellet, and (iii) their ability to grasp the pellet. Success rate was calculated by ratio of successful retrieval to number of attempts made.
Grip strength.
The grip strength test was used to evaluate forelimb muscular strength. The mice were placed on the testing apparatus, illuminated at 150 Lux, consisting of a metal grid attached to a dynamometer (Bioseb) that they naturally gripped onto and pulled back by their tail. Three consecutive trials were performed. The grip strength is calculated as a ratio of gripping force (grams) to the weight of the mouse (grams).
Notch bar.
The notch bar test was used to test hind limb coordination. The testing apparatus consists of a wooden bar (50-cm long) bearing 12 platforms (2 cm2) at regular gap intervals of 2 cm. The training was done on a flat wooden bar (without platforms) of similar dimensions. The mice were made to walk across the notched bar 20 times. The number of errors, defined by the hind limb missing the platform by slipping through the gap or off the platform, was calculated, and the global percentage of error was calculated.
DigiGait.
The mice were evaluated for ataxic behavior using the DigiGait imaging system (Mouse Specifics). The mice were placed on a treadmill controlled at a specific speed (18 cm/s), illuminated at 150 Lux, while a camera recorded their movements. The mice were made to run through the apparatus, and the analysis was made on a selected segment of 5 s. The software automatically detects the advance and retreat of each limb (strides), contact time with the treadmill (stance), and the intermediate period (swing). A total of 202 parameters associated with the postural and kinematic gait measurements, including time of movements, length of movements, braking and propelling, paw angle, etc. for each paw, as well as an “ataxic coefficient” were automatically generated by the software.
Hot plate.
We evaluated different aspects of sensory processing in the mice using three tests: hot plate test, adhesive removal test, and shock test. The hot plate test was used to evaluate thermal sensitivity. The mice were tested in two trials, where they were placed into a glass cylinder on a hot plate (Bioseb) set to 52 °C (illuminated at 115 Lux), and the latency to the first reaction (lick, flinch, shake, and jump) was recorded for a duration of 30 s in the first trial and 180 s in the second trial. The test was only made on hind paws.
Adhesive removal.
To evaluate touch sensitivity in the forepaws, we used the adhesive removal test. The test spanned across 5 d of habituation and 1 test day. Leukoplast (medical tape) was cut into 0.3 × 0.4-cm rectangles and carefully placed on the pads of right and left forepaws always in the same orientation, and the mice were placed in a cage without bedding illuminated at 150 Lux. The latency of the mouse to acknowledge the presence of the adhesive material (by sniffing, licking, or wiggling the limb), termed “contact time,” and latency to remove the adhesive, termed “removal time,” were recorded for each paw. For each animal and trial, four additional parameters were also collected: the time to touch and time to remove the adhesives tapes on both the left and right forepaws; 120 s was the maximum duration given to the mouse to remove the two adhesives, beyond which time the test was stopped.
Shock threshold.
We evaluated pain sensitivity using the shock threshold test. The mice were placed in a chamber (18.5 × 18 × 21.5 cm) illuminated at 150 Lux, where the floor is composed of stainless steel rods through which the foot shock is delivered (Coulbourn Instruments). The test was initiated by delivering a shock of 0.1 mA, and the intensity was gradually increased by 0.05 mA, with intervals of 30 s between two shocks, until a flinch is observed. Foot shocks were manually applied for 1 s, and behavioral responses were noted. The process continued until the mouse displayed vocalization and jumped, and the threshold for each reaction was recorded.
Social behavior.
Social interaction was evaluated by putting two mice of the same genotype, sex, and weight but from different cages in an open field (dimensions 44.3 × 44.3 × 16.8 cm) illuminated at 50 Lux for a duration of 10 min. We observed several parameters related to social behavior, such as sniffing, pawing, following, aggressiveness, and individual behavior, and recorded the time spent on each.
Social recognition.
The social recognition test is similar to the novel object recognition test, except that, instead of objects, we evaluate the tendency of a test mouse to explore an unfamiliar mouse placed in their proximity. The testing arena is made up of three identical chambers of identical size (illuminated at 90 Lux), a central chamber with two adjacent chambers connected internally with doors. In each adjacent chamber, a goal box was positioned delimited by a sliding grid. The test mouse was first allowed to freely explore the three chambers for 10 min. Next, the test mouse was placed in the central chamber, and an unfamiliar mouse was briefly introduced into one of the adjacent wire boxes kept closed. The doors of the central chamber were opened; the test mouse explored the three chambers, and its exploratory behavior was recorded for 10 min, specifically its preference for each of the wire boxes (with mouse or without mouse). Again, the test mouse was placed in the central chamber, and another unfamiliar mouse (novel mouse) was introduced briefly into the other wire box kept closed. The doors of the central chamber were opened; the test mouse explored the three chambers, and its exploratory behavior was recorded for 10 min. Its preference for exploring the wire box of the novel as opposed to the wire box of the familiar mouse was evaluated.
Methods for Yeast Work.
The WDR47 and LIS1 cDNAs inserted into the pENTR223 entry vector (ID: HsCD00505835 and ID: HsCD00515632; DNASU) were cloned by the Gateway (Invitrogen) method into pDONR221 entry vector and then recombined into yeast destination vectors (Addgene) (60) to obtain pAG413-promGPD-WDR47 (pSF358), pAG413- and pAG423-promGPD-WDR47-EGFP (pSF362 and pSF363), pAG413-promGPD-LIS1 (pSF356), and pAG413- and pAG423-promGPD-LIS1-EGFP (pSF360 and pSF361) plasmids. pAG413 is a low-copy number CEN plasmid, and pAG423 is a high-copy number 2-micron plasmid; both contain the HIS3 auxotrophic marker for selection of the transformants on SC-His medium. Plasmid sequences were verified (GATC Biotech). The promCUP1-mCherry-V5-ATG8 (pFL78, LEU2 selection marker) plasmid was a gift from Fulvio Reggiori (Center for Molecular Medicine, Utrecht, The Netherlands). The S. cerevisiae WT BY4742 (MATα leu2Δ0 ura3Δ0 his3Δ0 lys2Δ0) reference strain and the vps15∆ (MATα leu2Δ0 ura3Δ0 his3Δ0 lys2Δ0 vps15::KanMX) mutant strain were used. Yeast strains were grown at 30 °C to midexponential growth phase in rich yeast extract–peptone–dextrose (YPD) medium (1% yeast extract, 2% peptone, 2% glucose) in synthetic complete medium to maintain the plasmid SC-His [0.67% yeast nitrogen base (YNB) without amino acids, 2% glucose] and the appropriate His dropout mix or in autophagy induction medium SD-N (0.17% YNB without ammonium sulfate, without amino acids, 2% glucose). Yeast cells were transformed using the modified lithium acetate method (61).
For Western blot, total yeast protein extracts were obtained by NaOH lysis of 1.5 OD600 units of yeast cells followed by trichloroacetic acid precipitation, and pellet was resuspended in 50 μL of 2× Laemmli buffer plus Tris Base. Samples were incubated for 5 min at 37 °C before Western blot analysis by 8% SDS/PAGE followed by transfer on a nitrocellulose blotting membrane (Amersham Protran 0.45-μm NC) and immunoblotting with anti-GFP (1/1,000; 11814460001; Roche) antibodies using standard procedures. Images were acquired with the ChemiDoc Touch Imaging System (Bio-Rad). The BY4742 cells bearing the empty vector or the LIS1 or WDR47 expression vectors were grown at 30 °C to midexponential growth phase in SC-His medium, and 1 OD600 unit of cells was harvested by a 500 × g centrifugation for 1 min and resuspended in 50 µL YPD medium; vacuoles were stained with 2 µL FM4-64 (200 µM; Invitrogen) for 15 min at 30 °C before washing with 900 µL YPD and chasing by incubation at 30 °C for 10 min followed by a second wash in SC complete medium. The stained living yeast cells were observed by fluorescent microscopy.
Autophagy was analyzed on yeast cells expressing LIS1 or WDR47 (either nontagged from pSF356 or pSF358 plasmids or EGFP tagged from pSF360 or pSF362 plasmids) transformed with the mCherry-ATG8 expression vector (pFL78). Cells grown in CuSO4 (1 mM) containing SC-His-Leu medium to induce the expression of mCherry-ATG8 were harvested at OD600 0.5–1, washed with SD-N medium, resuspended in SD-N medium, and incubated at 30 °C for 4 h before observation by fluorescent microscopy. Observation was performed with 100×/1.45 oil objective (Zeiss) on a fluorescence Axio Observer D1 microscope (Zeiss) using GPF or DsRED filter and DIC optics. Images were captured with a CoolSnap HQ2 photometrix camera (Roper Scientific) and treated by ImageJ.
Yeast Two Hybrid.
Baits.
A cDNA fragment containing the two N-terminal domains (LisH and CTLH; amino acids 10–102) of WDR47 was PCR-amplified from the pEYFP-WDR47 (clone IOH26831; Imagines) using oligonucleotides WDR47-NdeI 5′ACTGCAGAACATATGAAAGAGGTTGAAATCATTAAG3′and WDR47-SalI 5′ACTGCAGAAGTCGACCTATGACATCGCGTTGTTA3′ and cloned in frame with the GAL4-DNA binding domain of the CLONTECH yeast two-hybrid (Y2H) bait vector, pGBKT7 (pGBKWDR47).
Preys.
A pretransformed MATCHMAKER library (CLONTECH), consisting of S. cerevisiae strain Y187, transformed with a fetal brain cDNA library was used for Y2H library screening.
Y2H screening.
pGBK-WDR47 was transformed into S. cerevisiae strain AH109 (CLONTECH) and mated with the S. cerevisiae Y187 pretransformed fetal brain cDNA library line. Diploid clones were grown on solid medium lacking Leu, Trp, and His for 10 d and then transferred to solid medium lacking these amino acids as well as Ade for another 8 d. Colonies positive for the reporter genes ADE2 and HIS3 were subsequently tested for expression of the reporter gene MEL1 by X-α-galactosidase filter assay. Colonies expressing all reporter genes were further analyzed. Prey plasmids from representative clones were rescued from diploid yeast cells via transformation of yeast DNA preparations into Escherichia coli DH5α and selection on ampicillin-containing media. These plasmids were retransformed into S. cerevisiae Y187 and tested for their ability to activate reporter genes in the presence of heterologous baits, including the GAL4-DNA binding domain alone or fused to murine p53 or cardiac myosin binding protein domain C5. Preys showing specific interaction were nucleotide sequenced, and the in-frame ORF sequences were analyzed (https://blast.ncbi.nlm.nih.gov/Blast.cgi) with BLASTN (against GenBank database) and BLASTP (against SwissProt database) to assign identity to the in-frame prey protein.
WDR47 Colocalization with SCG10.
GT1-7 cells were washed with PBS 24 h after transfection with pEYFP-WDR47, fixed with a 1:1 methanol:acetone solution and incubated for 10 min at 4 °C. This was followed by room temperature incubation in 5% donkey serum for 20 min. Cells were incubated for 90 min at room temperature with goat anti-SCG10 polyclonal antibody (Santa Cruz Biotechnology Inc). Cells were then washed three times with PBS and incubated for another 30 min with Texas Red-conjugated donkey anti-mouse secondary antibody (Santa Cruz Biotechnology Inc.); 10 mg/mL Hoechst 33342 (Sigma-Aldrich) was added in a 1:200 dilution for another 10-min room temperature incubation. Cover glasses were washed three times with PBS, transferred to microscope slides, and mounted with fluorescent mounting medium (Dako Cytomation).
Cell Culture and Transfection.
HEK293T.
HEK cells (HEK 293) used in Y2H were maintained at 37 °C and 5% CO2 in DMEM supplemented with 50% Ham’s F-12, 10% FBS, penicillin (100 U/mL), and streptomycin (100 mg/mL).
GT1-7.
Rat GT1-7 hypothalamus cells (provided by Pamela Mellon, University of California, San Diego, CA) used in live cell immunofluorescence colocalization were maintained at 37 °C and 5% CO2 in DMEM supplemented with 10% FCS, 4% glutamine, and 1% penicillin/streptomycin. Cells to be used for colocalization experiments were grown on sterile coverslips in six-well cell culture plates.
pEYFP-WDR47.
Clone IOH26831, which encodes the full-length WDR47 fused to an N-terminal enhanced YFP, and pCrl were transfected into GT1-7 and HEK293T cells using pCrl encoding full-length GeneJuice (Novagen) according to the manufacturer’s protocol.
Human Samples and WDR47 Mutation Screening.
Three cohorts of patients were specifically screened for WDR47 mutations. The first was a cohort of 50 patients with unexplained lissencephaly. The second was a cohort of 30 patients with nonsyndromic corpus callosum agenesis (62). The third was a cohort of 207 patients with undiagnosed intellectual disability (environmental causes, excluded and negative for Fragile-X testing). In all cases, a comparative genomic hybridization array was achieved to exclude the presence of copy number variants. For the first two cohorts, we screened WDR47 for mutation by Sanger sequencing. We designed primers along the longest WDR47 transcript to target all possible coding regions. Primers were designed to target and amplify 15 exons (Dataset S13). The PCR mix consisted of a 25 µL reaction made of 1.25 µL DNA (5 ng/µL), 2.5 µL 10× PCR buffer (without magnesium), 0.75 µL 50 mM MgCl2, 0.25 µL 10 mM dNTP, 19 µL H2O, 0.25 µL Platinum Taq (10966–034; Invitrogen), 0.5 µL 10 mM forward primer, and 0.5 µL 10 mM reverse primer. PCR consisted of an activation of Platinum Taq enzyme at 94 °C for 5 min (1 cycle), DNA denaturation at 94 °C for 30 s, annealing of primers to DNA strands at 60 °C for 30 s, extension of primers at 72 °C for 45 s (35 cycles), and a final extension at 72 °C for 5 min (1 cycle). A touch down program was used when the classical PCR did not work. The amplicons were sent to GATC or Eurofin for purification and sequencing. Sequences were analyzed using Seqscape to identify synonymous or nonsynonymous mutations, splice acceptor/donor site mutations, and short coding insertion/deletions. To minimize false negative rate, we set up a het signal threshold of at least 28%. For the third cohort, mutation screening was performed using a targeted high-throughput sequencing approach: WDR47 was included in a list of 275 confirmed or candidate ID genes. Library preparation, sequencing, and variant analysis were performed as previously described (63). When required, we designed sets of primers to validate the mutations found (Dataset S13). The mutations were assessed for its deleteriousness using Polyphen (genetics.bwh.harvard.edu/pph2/) and SIFT (sift.jcvi.org/). Uncommon benign mutations observed in healthy individuals were excluded using 1000 Genome Project, dbSNP, Exome Variant Server, and Exome Aggregation Consortium.
Supplementary Material
Acknowledgments
We thank Jonathan Flint for helpful comments on the manuscript, Christel Depienne, and Albert Weixlbaumer for scientific discussions. We thank Sylvie Jacquot at the Mouse Clinical Institute (MCI) for the development of the genotyping strategy. We thank Nadia Messadeq at the Electron Microscopy Facility and Mustapha Oulad-Abdelghani for producing WDR47 antibody. We also thank animal care workers at the MCI and Marc Koch at Bruker Nano Surfaces Division for his help with acquiring the superresolution images. We thank Hugues Jacobs at the MCI for advice on histology and all of the members of the imaging facility at Institut de Génétique et de Biologie Moléculaire et Cellulaire. We thank Fulvio Reggiori (Utrecht University) for sharing mCherry-Atg8 plasmid and Gabriele Grenningloh (École Polytechnique Fédérale de Lausanne) for SCG10 antibodies. J.D.G. is funded by the Action Thématique et Incitative sur Programme (ATIP)–Avenir joint CNRS-INSERM program and the Fyssen foundation. This study was funded by CNRS (S.F.), INSERM (E.B. and S.F.), Strasbourg University (S.F.), Initiatives d'Excellence (IDEX) 2015 Attractivité (S.B.), and Grant ANR-10-LABX-0030-INRT, a French State fund managed by the Agence Nationale de la Recherche under the frame program Investissements d'Avenir ANR-10-IDEX-0002-02 (to B.Y. and J.D.G.). C.K. is supported by funding from the South African Medical Research Council. B.Y. is supported by the Jerôme Lejeune Foundation, the French National Research Agency (ANR-11-PDOC-0029), and the Gutenberg Circle.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
2Sanger Mouse Genetics Project: Valerie E. Vancollie, Lauren F. E. Anthony, Simon A. Maguire, David Lafont, Selina A. Pearson, Amy S. Gates, Mark Sanderson, Carl Shannon, Maksymilian T. Sumowski, Robbie S. B. McLaren-Jones, Agnieszka Swiatkowska, Christopher M. Isherwood, Emma L. Cambridge, Heather M. Wilson, Susana S. Caetano, Anna Karin B. Maguire, Antonella Galli, Anneliese O. Speak, Joshua Dench, Elizabeth Tuck, Jeanne Estabel, Angela Green, Catherine Tudor, Emma Siragher, Monika Dabrowska, Cecilia Icoresi Mazzeo, Yvette Hooks, Fiona Kussy, Mark Griffiths, David Gannon, Brendan Doe, Katharina Boroviak, Hannah Wardle-Jones, Nicola Griggs, Joanna Bottomley, Ed Ryder, Diane Gleeson, Jacqueline K. White, Ramiro Ramirez-Solis, and Christopher J. Lelliott.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713625114/-/DCSupplemental.
Contributor Information
Collaborators: Valerie E. Vancollie, Lauren F. E. Anthony, Simon A. Maguire, David Lafont, Selina A. Pearson, Amy S. Gates, Mark Sanderson, Carl Shannon, Maksymilian T. Sumowski, Robbie S. B. McLaren-Jones, Agnieszka Swiatkowska, Christopher M. Isherwood, Emma L. Cambridge, Heather M. Wilson, Susana S. Caetano, Anna Karin B. Maguire, Antonella Galli, Anneliese O. Speak, Joshua Dench, Elizabeth Tuck, Jeanne Estabel, Angela Green, Catherine Tudor, Emma Siragher, Monika Dabrowska, Cecilia Icoresi Mazzeo, Yvette Hooks, Fiona Kussy, Mark Griffiths, David Gannon, Brendan Doe, Katharina Boroviak, Hannah Wardle-Jones, Nicola Griggs, Joanna Bottomley, Ed Ryder, Diane Gleeson, Jacqueline K. White, Ramiro Ramirez-Solis, and Christopher J. Lelliott
References
- 1.Andrade MA, Perez-Iratxeta C, Ponting CP. Protein repeats: Structures, functions, and evolution. J Struct Biol. 2001;134:117–131. doi: 10.1006/jsbi.2001.4392. [DOI] [PubMed] [Google Scholar]
- 2.Sondek J, Bohm A, Lambright DG, Hamm HE, Sigler PB. Crystal structure of a G-protein beta gamma dimer at 2.1A resolution. Nature. 1996;379:369–374. doi: 10.1038/379369a0. [DOI] [PubMed] [Google Scholar]
- 3.Stirnimann CU, Petsalaki E, Russell RB, Müller CW. WD40 proteins propel cellular networks. Trends Biochem Sci. 2010;35:565–574. doi: 10.1016/j.tibs.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Smith TF, Gaitatzes C, Saxena K, Neer EJ. The WD repeat: A common architecture for diverse functions. Trends Biochem Sci. 1999;24:181–185. doi: 10.1016/s0968-0004(99)01384-5. [DOI] [PubMed] [Google Scholar]
- 5.Hurtley S. Spatial cell biology. Location, location, location. Introduction. Science. 2009;326:1205. doi: 10.1126/science.326.5957.1205. [DOI] [PubMed] [Google Scholar]
- 6.Sasaki S, et al. A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron. 2000;28:681–696. doi: 10.1016/s0896-6273(00)00146-x. [DOI] [PubMed] [Google Scholar]
- 7.Saillour Y, et al. LIS1-related isolated lissencephaly: Spectrum of mutations and relationships with malformation severity. Arch Neurol. 2009;66:1007–1015. doi: 10.1001/archneurol.2009.149. [DOI] [PubMed] [Google Scholar]
- 8.Bilgüvar K, et al. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature. 2010;467:207–210. doi: 10.1038/nature09327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colin E, et al. Loss-of-function mutations in WDR73 are responsible for microcephaly and steroid-resistant nephrotic syndrome: Galloway-Mowat syndrome. Am J Hum Genet. 2014;95:637–648. doi: 10.1016/j.ajhg.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindwall C, Fothergill T, Richards LJ. Commissure formation in the mammalian forebrain. Curr Opin Neurobiol. 2007;17:3–14. doi: 10.1016/j.conb.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 11.Paul LK, et al. Agenesis of the corpus callosum: Genetic, developmental and functional aspects of connectivity. Nat Rev Neurosci. 2007;8:287–299. doi: 10.1038/nrn2107. [DOI] [PubMed] [Google Scholar]
- 12.Paul LK, Corsello C, Kennedy DP, Adolphs R. Agenesis of the corpus callosum and autism: A comprehensive comparison. Brain. 2014;137:1813–1829. doi: 10.1093/brain/awu070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, et al. A comparative diffusion tensor imaging study of corpus callosum subregion integrity in bipolar disorder and schizophrenia. Psychiatry Res. 2014;221:58–62. doi: 10.1016/j.pscychresns.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 14.Balevich EC, et al. Corpus callosum size and diffusion tensor anisotropy in adolescents and adults with schizophrenia. Psychiatry Res. 2015;231:244–251. doi: 10.1016/j.pscychresns.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edwards TJ, Sherr EH, Barkovich AJ, Richards LJ. Clinical, genetic and imaging findings identify new causes for corpus callosum development syndromes. Brain. 2014;137:1579–1613. doi: 10.1093/brain/awt358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chédotal A. Further tales of the midline. Curr Opin Neurobiol. 2011;21:68–75. doi: 10.1016/j.conb.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 17.Dent EW, Gupton SL, Gertler FB. The growth cone cytoskeleton in axon outgrowth and guidance. Cold Spring Harb Perspect Biol. 2011;3:a001800. doi: 10.1101/cshperspect.a001800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lowery LA, Van Vactor D. The trip of the tip: Understanding the growth cone machinery. Nat Rev Mol Cell Biol. 2009;10:332–343. doi: 10.1038/nrm2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sapir T, Elbaum M, Reiner O. Reduction of microtubule catastrophe events by LIS1, platelet-activating factor acetylhydrolase subunit. EMBO J. 1997;16:6977–6984. doi: 10.1093/emboj/16.23.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen JF, et al. Microcephaly disease gene Wdr62 regulates mitotic progression of embryonic neural stem cells and brain size. Nat Commun. 2014;5:3885. doi: 10.1038/ncomms4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirschner MW, Mitchison T. Microtubule dynamics. Nature. 1986;324:621. doi: 10.1038/324621a0. [DOI] [PubMed] [Google Scholar]
- 22.Richards LJ, Plachez C, Ren T. Mechanisms regulating the development of the corpus callosum and its agenesis in mouse and human. Clin Genet. 2004;66:276–289. doi: 10.1111/j.1399-0004.2004.00354.x. [DOI] [PubMed] [Google Scholar]
- 23.Wang W, et al. Nemitin, a novel Map8/Map1s interacting protein with Wd40 repeats. PLoS One. 2012;7:e33094. doi: 10.1371/journal.pone.0033094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Llewellyn KJ, et al. Lipid-enriched diet rescues lethality and slows down progression in a murine model of VCP-associated disease. Hum Mol Genet. 2014;23:1333–1344. doi: 10.1093/hmg/ddt523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan Q, et al. Folic acid supplementation changes the fate of neural progenitors in mouse embryos of hyperglycemic and diabetic pregnancy. J Nutr Biochem. 2013;24:1202–1212. doi: 10.1016/j.jnutbio.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 26.Brizuela M, et al. The microtubule-stabilizing drug Epothilone D increases axonal sprouting following transection injury in vitro. Mol Cell Neurosci. 2015;66:129–140. doi: 10.1016/j.mcn.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 27.Wu CL, et al. Interplay between cell migration and neurite outgrowth determines SH2B1β-enhanced neurite regeneration of differentiated PC12 cells. PLoS One. 2012;7:e34999. doi: 10.1371/journal.pone.0034999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang CC, Park AY, Guan JL. In vitro scratch assay: A convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc. 2007;2:329–333. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- 29.Riederer BM, et al. Regulation of microtubule dynamics by the neuronal growth-associated protein SCG10. Proc Natl Acad Sci USA. 1997;94:741–745. doi: 10.1073/pnas.94.2.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tararuk T, et al. JNK1 phosphorylation of SCG10 determines microtubule dynamics and axodendritic length. J Cell Biol. 2006;173:265–277. doi: 10.1083/jcb.200511055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bondallaz P, Barbier A, Soehrman S, Grenningloh G, Riederer BM. The control of microtubule stability in vitro and in transfected cells by MAP1B and SCG10. Cell Motil Cytoskeleton. 2006;63:681–695. doi: 10.1002/cm.20154. [DOI] [PubMed] [Google Scholar]
- 32.Tucci V, et al. Reaching and grasping phenotypes in the mouse (Mus musculus): A characterization of inbred strains and mutant lines. Neuroscience. 2007;147:573–582. doi: 10.1016/j.neuroscience.2007.04.034. [DOI] [PubMed] [Google Scholar]
- 33.Bouet V, et al. The adhesive removal test: A sensitive method to assess sensorimotor deficits in mice. Nat Protoc. 2009;4:1560–1564. doi: 10.1038/nprot.2009.125. [DOI] [PubMed] [Google Scholar]
- 34.Zhang C, Zhang F. The multifunctions of WD40 proteins in genome integrity and cell cycle progression. J Genomics. 2015;3:40–50. doi: 10.7150/jgen.11015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stoetzel C, et al. A mutation in VPS15 (PIK3R4) causes a ciliopathy and affects IFT20 release from the cis-Golgi. Nat Commun. 2016;7:13586. doi: 10.1038/ncomms13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emes RD, Ponting CP. A new sequence motif linking lissencephaly, Treacher Collins and oral-facial-digital type 1 syndromes, microtubule dynamics and cell migration. Hum Mol Genet. 2001;10:2813–2820. doi: 10.1093/hmg/10.24.2813. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki SW, Onodera J, Ohsumi Y. Starvation induced cell death in autophagy-defective yeast mutants is caused by mitochondria dysfunction. PLoS One. 2011;6:e17412. doi: 10.1371/journal.pone.0017412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shibutani ST, Yoshimori T. A current perspective of autophagosome biogenesis. Cell Res. 2014;24:58–68. doi: 10.1038/cr.2013.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kabeya Y, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen LS, et al. A nonsense variant in HERC1 is associated with intellectual disability, megalencephaly, thick corpus callosum and cerebellar atrophy. Eur J Hum Genet. 2016;24:455–458. doi: 10.1038/ejhg.2015.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grenningloh G, Soehrman S, Bondallaz P, Ruchti E, Cadas H. Role of the microtubule destabilizing proteins SCG10 and stathmin in neuronal growth. J Neurobiol. 2004;58:60–69. doi: 10.1002/neu.10279. [DOI] [PubMed] [Google Scholar]
- 42.Komulainen E, et al. JNK1 controls dendritic field size in L2/3 and L5 of the motor cortex, constrains soma size, and influences fine motor coordination. Front Cell Neurosci. 2014;8:272. doi: 10.3389/fncel.2014.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mackeh R, Perdiz D, Lorin S, Codogno P, Poüs C. Autophagy and microtubules–New story, old players. J Cell Sci. 2013;126:1071–1080. doi: 10.1242/jcs.115626. [DOI] [PubMed] [Google Scholar]
- 44.Monastyrska I, Rieter E, Klionsky DJ, Reggiori F. Multiple roles of the cytoskeleton in autophagy. Biol Rev Camb Philos Soc. 2009;84:431–448. doi: 10.1111/j.1469-185X.2009.00082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie R, Nguyen S, McKeehan WL, Liu L. Acetylated microtubules are required for fusion of autophagosomes with lysosomes. BMC Cell Biol. 2010;11:89. doi: 10.1186/1471-2121-11-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu HJ, Pu JL, Krafft PR, Zhang JM, Chen S. The molecular mechanisms between autophagy and apoptosis: Potential role in central nervous system disorders. Cell Mol Neurobiol. 2015;35:85–99. doi: 10.1007/s10571-014-0116-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dragich JM, et al. Autophagy linked FYVE (Alfy/WDFY3) is required for establishing neuronal connectivity in the mammalian brain. Elife. 2016;5:e14810. doi: 10.7554/eLife.14810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lek M, et al. Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuma A, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 50.Cahana A, et al. Targeted mutagenesis of Lis1 disrupts cortical development and LIS1 homodimerization. Proc Natl Acad Sci USA. 2001;98:6429–6434. doi: 10.1073/pnas.101122598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dujardin DL, et al. A role for cytoplasmic dynein and LIS1 in directed cell movement. J Cell Biol. 2003;163:1205–1211. doi: 10.1083/jcb.200310097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kobayashi N, et al. RanBPM, Muskelin, p48EMLP, p44CTLH, and the armadillo-repeat proteins ARMC8alpha and ARMC8beta are components of the CTLH complex. Gene. 2007;396:236–247. doi: 10.1016/j.gene.2007.02.032. [DOI] [PubMed] [Google Scholar]
- 53.Alibhoy AA, Giardina BJ, Dunton DD, Chiang HL. Vid30 is required for the association of Vid vesicles and actin patches in the vacuole import and degradation pathway. Autophagy. 2012;8:29–46. doi: 10.4161/auto.8.1.18104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang B, et al. The microtubule-stabilizing agent, epothilone D, reduces axonal dysfunction, neurotoxicity, cognitive deficits, and Alzheimer-like pathology in an interventional study with aged tau transgenic mice. J Neurosci. 2012;32:3601–3611. doi: 10.1523/JNEUROSCI.4922-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Skarnes WC, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karp NA, Melvin D, Mott RF. Sanger Mouse Genetics Project Robust and sensitive analysis of mouse knockout phenotypes. PLoS One. 2012;7:e52410. doi: 10.1371/journal.pone.0052410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wahlsten D, Metten P, Crabbe JC. Survey of 21 inbred mouse strains in two laboratories reveals that BTBR T/+ tf/tf has severely reduced hippocampal commissure and absent corpus callosum. Brain Res. 2003;971:47–54. doi: 10.1016/s0006-8993(03)02354-0. [DOI] [PubMed] [Google Scholar]
- 58.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 59.Mikhaleva A, Kannan M, Wagner C, Yalcin B. Histomorphological phenotyping of the adult mouse brain. Curr Protoc Mouse Biol. 2016;6:307–332. doi: 10.1002/cpmo.12. [DOI] [PubMed] [Google Scholar]
- 60.Alberti S, Gitler AD, Lindquist S. A suite of gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast. 2007;24:913–919. doi: 10.1002/yea.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gietz D, St Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Romaniello R, et al. Clinical characterization, genetics, and long-term follow-up of a large cohort of patients with agenesis of the corpus callosum. J Child Neurol. 2016;32:60–71. doi: 10.1177/0883073816664668. [DOI] [PubMed] [Google Scholar]
- 63.Redin C, et al. Efficient strategy for the molecular diagnosis of intellectual disability using targeted high-throughput sequencing. J Med Genet. 2014;51:724–736. doi: 10.1136/jmedgenet-2014-102554. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.