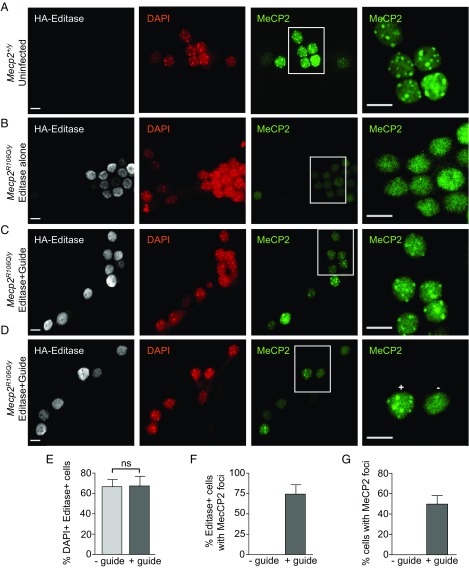
Fig. 5.
Site-directed RNA editing restores the ability of MeCP2 to bind to heterochromatin. Shown are representative confocal images of hippocampal neurons (DIV14) immunolabeled for Editase (HA) and MeCP2. DAPI staining outlines the nuclei and shows heterochromatic foci. Insets demarcate the cells imaged at higher magnification and higher gain in the adjacent panels. (A) Wild-type (Mecp2+/y) neuronal cultures. (B) Mecp2R106Q/y neuronal cultures transduced with AAV1/2 virus expressing Editase alone (no guide). These neurons never exhibited MeCP2 enrichment in heterochromatin. (C and D) Mecp2R106Q/y neuronal cultures transduced with AAV1/2 virus expressing Editase and guide containing the C mismatch at the target A. In D, + and − indicate nuclei with the presence and absence, respectively, of MeCP2 enrichment in heterochromatin. While there appears to be a relationship between Editase/guide expression and MeCP2 heterochromatic enrichment in this image, overall we were not able to confidently support this relationship. (Scale bar, 10 μm.) (E and G) Each histogram represents quantification of cells (Editase alone, n = 134; Editase and guide, n = 137) from three fields in each of three slides (mean ± SD). (E) Percentage of Editase+ cells identified by HA nuclear staining after thresholding signals from uninfected cells. Percentages are relative to the total number of DAPI+ cells. (F) Percentage of Editase+ cells with MeCP2 enrichment in heterochromatin (foci) and (G) percentage of all cells with MeCP2 enrichment in heterochromatin (foci). ns, not significant.