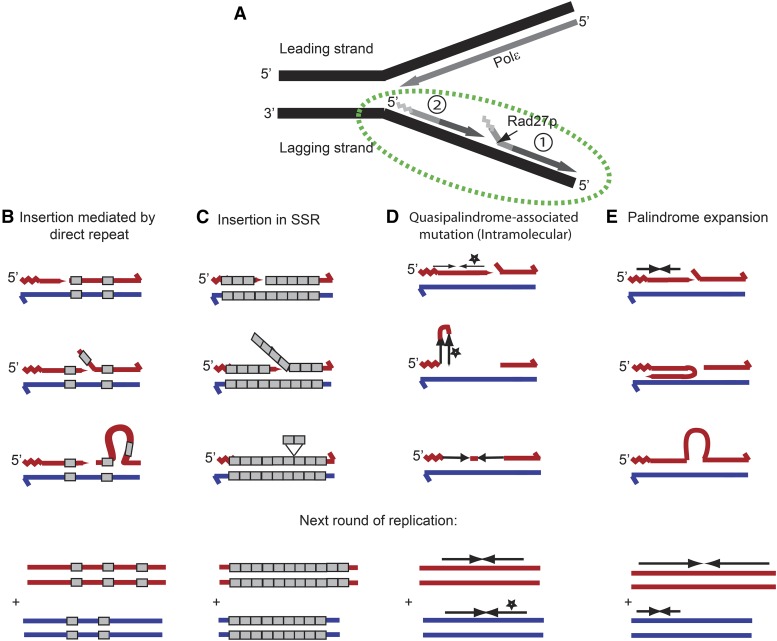
Figure 10.
Models for the formation of the common events observed in rad27Δ strains. (A) Description of Okazaki fragment maturation. Primers produced by polymerase α are shown in a light gray zigzag, and the extension by polymerase δ is represented by a darker shade of gray. The predicted position where Rad27/FEN1 acts to remove the RNA/DNA primer in wild-type strains is indicated by the small black arrow. The junction between two adjacent Okazaki fragments is enlarged in B–E (green ellipse). (B) A model for an insertion mediated by a direct repeat, where realignment of the direct repeat in the wrong location enables the formation of a loop containing the unique sequence that forms the insertion in the next round of replication. (C) A model for an insertion of one repeat in SSR. We note that mechanisms B and C are similar, as the repeat can easily realign in multiple places. (D) An intramolecular template-switching model for the correction of a quasi-palindrome to a palindrome. The star indicates a change between the two copies of the IR. (E) A model for expansion of a palindrome through a template-switching mechanism. We suggest that the polymerase is stalled due to stabilization of the flap as a hairpin in the previously synthesized Okazaki fragment, or due to inefficient strand displacement. The polymerase continues synthesis using the nascent strand as a template. The red strand is the nascent strand and the blue strand is the original template strand.