(
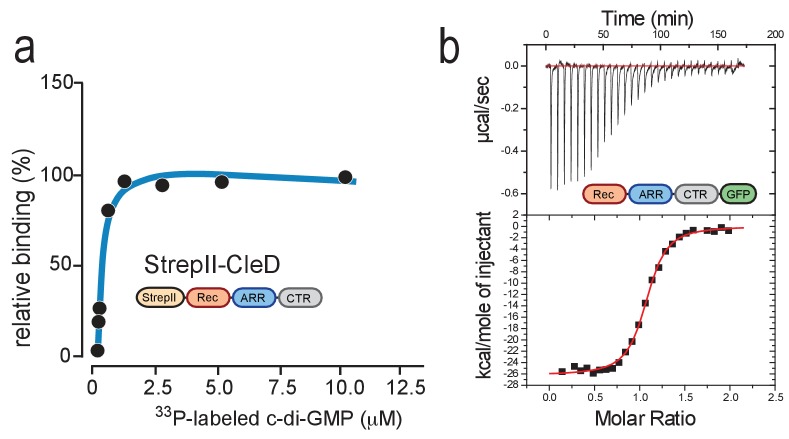
a) CleD binds c-di-GMP specifically. Purified StrepII-CleD (1 µM) was UV cross-linked in the presence of 0.25 µM
33P-labelled c-di-GMP and the indicated concentrations of non-labeled nucleotides. Binding is shown as relative units determined in the absence of non-labeled nucleotides. While equimolar amounts of non-labeled c-di-GMP reduced binding of
33P-labelled c-di-GMP, a similar reduction required 1000-fold higher concentrations of GTP or pGpG. (
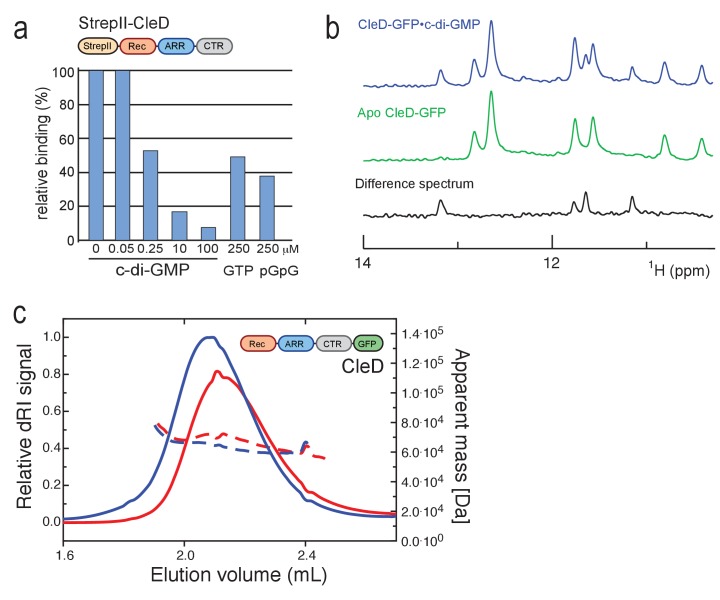
b) Excerpts of the one-dimensional
1H-NMR Jump-return spectra of CleD-GFP in the presence of c-di-GMP (blue), without c-di-GMP (green), and the difference spectrum of both. Four different imino H1 proton resonances are observed in the difference spectrum, indicative of four guanosines in an asymmetric environment. This is consistent with the binding of one c-di-GMP dimer to one protein monomer. A similar spectrum was observed for PA4608·c-di-GMP, which binds c-di-GMP as an intercalated dimer (
Habazettl et al., 2011). (
c) CleD is a monomer in its apo- and ligand bound form. SEC-MALS chromatograms of His-CleD-GFP in the absence (red) and presence of c-di-GMP (blue). Loading concentrations were 38.6 µM for CleD-GFP and 192.8 µM for c-di-GMP. Continuous lines represent the dRI signal (left axis) and broken lines the MALS derived apparent mass values (right axis). The calculated mass using ProtParam was 58.4 kDa for CleD-GFP and apparent masses are 69.2 kDa and 64.5 kDa for CleD-GFP and CleD-GFP/c-di-GMP, respectively.