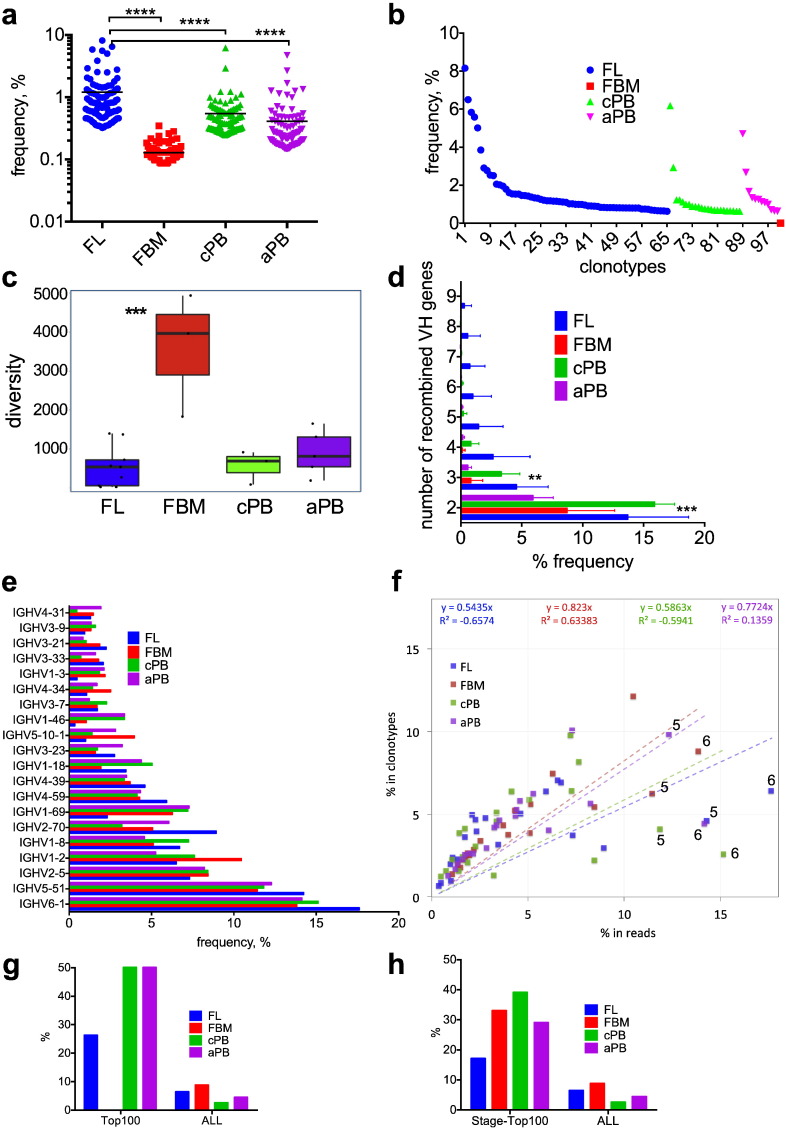
Fig. 2.
Clonotypic abundance and diversity in fetal and post-natal B-cells. a. Frequency (abundance counted in reads) of the 100 most abundant clonotypes in each developmental stage (horizontal lines indicate mean values, ****p < 0.0001). b. Distribution of the 100 most abundant clonotypes across the 4 developmental stages. c. Clonotypic diversity in each developmental stage as assessed by the inverse Simpson concentration (see Methods). ***p < 0.001 for FBM as the most diverse. d. Frequency of CDR3 peptides generated by convergent recombination of 2–9 different IGHV genes in each developmental stage. ** and *** for p < 0.01 and p < 0.001 respectively for FL and cPB vs. FBM and aPB. e. IGHV gene repertoire counted in reads in fetal and postnatal B-cells. Mean values are shown (error bars omitted for simplicity). f. Correlation of the relative clonotype abundances of the 20 most popular IGHV genes across developmental stages with their corresponding relative read counts. Lower slopes (as indicated by ‘y’ values in respective colors) of the regression lines for FL and cPB indicate the predominance of high abundance clonotypes in these developmental stages. ‘5’ and ‘6’: outlying and highly expressed IGHV5-51 and IGHV6-1 respectively. g & h. IGHV6-1 usage in the 100 most abundant clonotypes across developmental stages (g), and in the 100 most abundant clonotypes in each developmental stage (h); these “Top100” frequencies are compared to those of IGHV6-1 usage in all clonotypes (“ALL”).