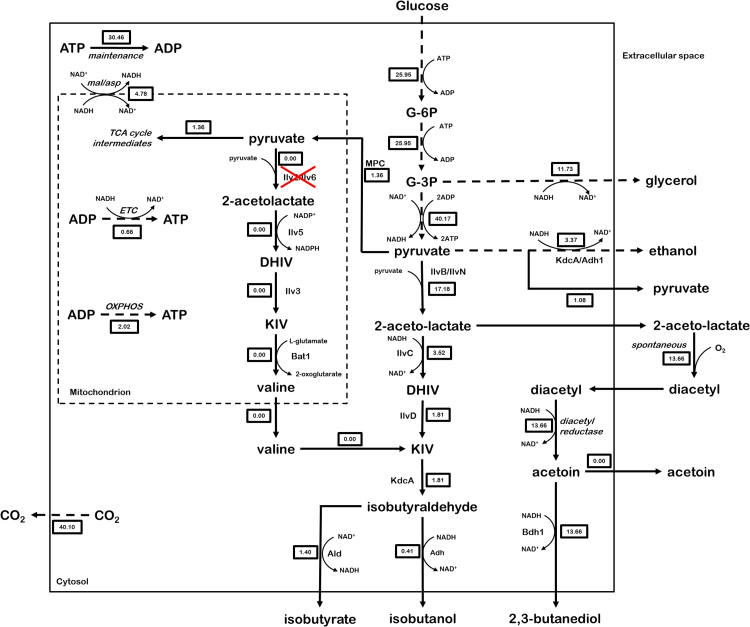
Fig. 5.
Flux distribution maps for S. cerevisiae IME307 (Δpdc1,5,6 Δilv2 MTH1ΔTcoilvBCDNcokdcA) grown in micro-aerobic cultures (see Table 6), calculated using CellNetAnalyzer. Dashed arrows represent multiple enzyme-catalysed reactions. Numbered boxes represent the modelled metabolic flux through each reaction (expressed in µmol/g biomass/h). Mal/asp: malate/aspartate shuttle, ETC: Electron transport chain, OXPHOS: Oxidative phosphorylation, G-6P: glucose-6-phosphate, G-3P: glyceraldehyde-3-phosphate, DHIV: 2,3-dihydroxyisovalerate, KIV: α-ketoisovalerate.