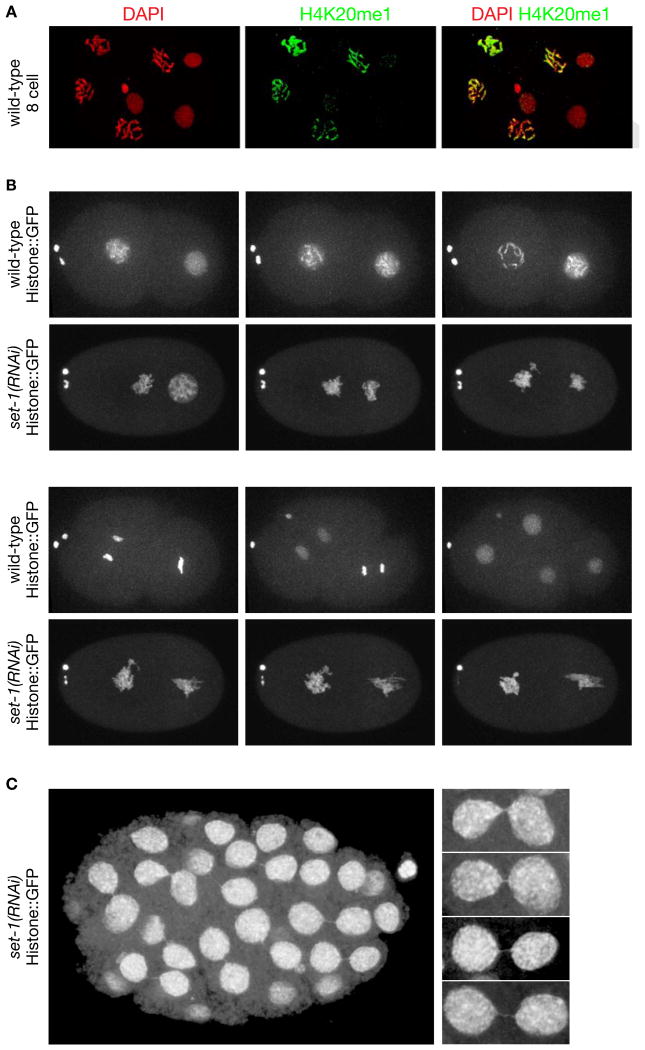
Figure 4. Depletion of Histone Methyl Transferase set−1 Causes Severe Mitotic Chromosome Segregation Defects.
(A) Confocal images of a wild-type 8-cell embryo stained with DAPI and H4K20me1 antibody. H4K20me1 is significantly elevated in mitotic vs. interphase (arrows) nuclei and is distributed across all chromosomes.
(B), (C) Live images of set−1(RNAi) embryos show severe mitotic defects. (B) Defective mitotic chromosome segregation caused embryonic lethality. The time series of chromosome segregation starts with 2-cell wild-type or set−1(RNAi) embryos expressing Histone::GFP. Both wild-type cells completed a round of division in 25 minutes. The set−1(RNAi) embryo arrested in mitosis and died. Arrow, two polar bodies. (C) Live image of an older set−1(RNAi) embryo that received less set−1 RNA than in (B) shows mitotic segregation defects in nearly all cells, including chromatin bridges and micronuclei (arrows). Enlargements of chromatin bridges are shown. Scale bars (A-C), 5 μm. See Table S2.