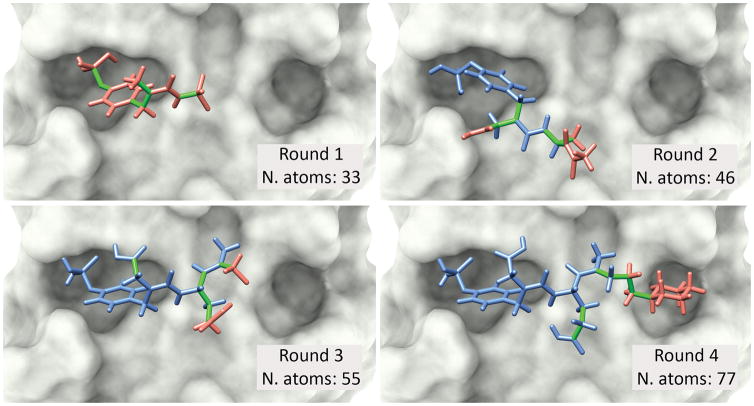
Figure 1. Incremental docking of a modified peptide.
The proto-oncogene tyrosine protein kinase Src (c-Src) has been associated with breast cancer and osteoporosis [11]. A known peptidomimetic inhibitor, capable of binding to the Src SH2 domain, has 77 atoms and 17 DoFs. This modified peptide provides a good example of the use of DINC’s incremental approach for binding mode prediction. DINC starts by selecting a small fragment of the ligand (top left), with only 6 DoFs (depicted in green), and using it as input for the first round of docking to the SH2 domain (depicted in grey). The best binding modes are selected across multiple parallel docking runs, and the corresponding fragments are expanded by adding a small number of atoms. These extended fragments are used as input for the second round of docking, in which a new subset of 6 flexible DoFs is explored. These flexible DoFs involve some of the “new” atoms (red) and some of the “old” atoms (blue), as depicted in the best binding mode obtained in round 2 (top right). This process continues until the entire ligand has been reconstructed (bottom left and right). In this example, the root mean square deviation (RMSD) between the obtained model and the corresponding crystal structure of the same complex (PDB code 1SKJ) is only 1.97 Å.