Figure 4.
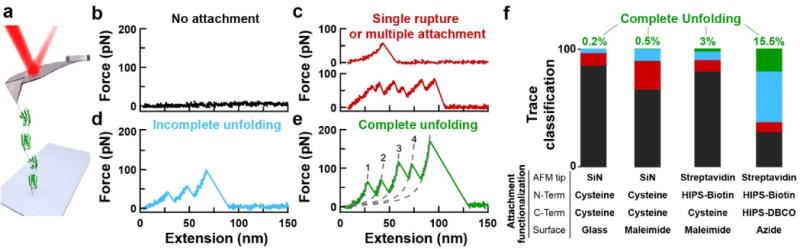
Improved single-molecule force spectroscopy. (a) A schematic of the experiment shows a polyprotein with four NuG2 domains being stretched between the surface and an AFM tip. (b–e) Force-extension curves (FECs) typically show one of four classes of mechanical fingerprints ranging from no attachment to full unfolding of the polyprotein. In panel e, the segments of the FEC between domain ruptures are well described by a worm-like-chain model (dashed lines). FEC data smoothed to 1 kHz. (f) Bar graphs characterizing the fraction of records with no unfolding (black), traces showing a single unfolding event or pulling on multiple proteins in parallel (red), incomplete unfolding of a single polypeptide (blue), and complete unfolding (green). Sitespecific attachment to the functionalized, PEG-coated surface used either a maleimide-thiol reaction or a reaction between dibenzocyclooctyne (DBCO), a copperless-click-chemistry reagent, and an azide-derivatized surface. Hydrazino-Pictet-Spengler (HIPS) reagents labelled genetically encoded aldehydes with biotin and DBCO. Going from left to right, the bar graphs were based on 595, 595, 1237, and 898 individual stretching attempts. SiN, silicon nitride.
